| Issue |
A&A
Volume 681, January 2024
|
|
|---|---|---|
| Article Number | A93 | |
| Number of page(s) | 9 | |
| Section | Numerical methods and codes | |
| DOI | https://doi.org/10.1051/0004-6361/202347651 | |
| Published online | 23 January 2024 | |
Adaptive data reduction workflows for astronomy: The ESO Data Processing System (EDPS)
European Southern Observatory,
Karl-Schwarzschild-Str. 2,
85748
Garching, Germany
e-mail: wfreudli@eso.org
Received:
3
August
2023
Accepted:
3
November
2023
Context. Astronomical data reduction is usually done via processing pipelines that consist of a series of individual processing steps that can be executed one at a time. These processing steps are then strung together into workflows and fed with data to address a particular processing goal. Examples of such pipeline processing goals are the quality control of incoming data from telescopes, unsupervised production of science and calibration products for an archive, and supervised data reduction to serve the specific science goals of a scientist. For each of these goals, individual workflows need to be developed. These workflows need to evolve when the pipeline, observing strategies, or calibration plans change. Writing and maintaining such a collection of workflows is therefore a complex and expensive task.
Aims. In this paper we propose a data processing system that automatically derives processing workflows for different use cases from a single specification of a cascade of processing steps.
Methods. The system works by using formalised descriptions of data processing pipelines that specify the input and output of each processing step. Inputs can be existing data or the output of a previous step. The rules for selecting the most appropriate input data are directly attached to the description.
Results. A version of the proposed system has been implemented as the ESO Data Processing System (EDPS) in the Python language. The specification of processing cascades and data organisation rules use a restrictive set of Python classes, attributes, and functions.
Conclusions. The EDPS implementation of the proposed system was used to demonstrate that it is possible to automatically derive from a single specification of a pipeline processing cascade the workflows that the European Southern Observatory uses for quality control, archive production, and specialised science reduction. The EDPS will be used to replace all data reduction systems using different workflow specifications that are currently in use at the European Southern Observatory.
Key words: methods: data analysis / methods: numerical / techniques: image processing / techniques: miscellaneous / virtual observatory tools
© The Authors 2024
 Open Access article, published by EDP Sciences, under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Open Access article, published by EDP Sciences, under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article is published in open access under the Subscribe to Open model. Subscribe to A&A to support open access publication.
1 Introduction
Modern astronomical telescopes and their instrumentation produce raw data that record the scientific signal, but also include the signature of the atmosphere, telescope, instrument, and detector(s). In addition, the raw data include noise from different sources. Data reduction is a necessary step before any data can be scientifically analysed and interpreted. Observatories such as the European Southern Observatory process data from their telescopes and distribute processed data through open archives to their user communities (e.g. Romaniello et al. 2023; Tran et al. 2020; Brodheim et al. 2022; Lacy 2016). At the same time, the end users of the data are often in the best position to carry out data reduction in a way that optimises the quality of the reduction for a particular purpose.
In addition to the data reduction for scientific use, processing the raw data also plays an important role in monitoring the quality of incoming data. A rigorous quality control process can detect hardware and software issues, and can monitor the impact of the ambient conditions on the data. At ESO observations go through three levels of quality control. The first level is done right after an observation is completed during the course of the night. This is followed by two off-line quality control processes, one that focuses on the data quality of one night and the characterisation of the observing system performance, and another that investigates the long-term quality of the science data. In this paper we present the design of a data reduction system that automatically adapts to such different use cases for data reduction, and automatically derives data reduction workflows for each of them.
2 Pipelines and data processing workflows
2.1 Basic concepts
With the ever-increasing complexity of astronomical instrumentation, it is inevitable that the data processing is a multi-step process that requires using highly specialised software tools. Observatories have long recognised the need to provide data processing pipelines (e.g. Weilbacher et al. 2020; Lemoine-Busserolle et al. 2019; Lockhart et al. 2019; Law et al. 2016; Lacy et al. 2015) to support the exploitation of their data. These pipelines typically consist of a series of stand-alone modules (‘recipes’) that have to be applied in sequence to the data. Each recipe is usually designed to process a single data type as its principal input. Associated calibration files are used in the processing of these input files. The algorithms used by the recipes usually have some parameters that can be set at execution time.
The input files of a recipe can either be unprocessed raw data, the output from previous processing steps, or pre-computed products called static calibrations. These recipes need to be combined into a data processing workflow that specifies the sequence of processing steps to process a pool of data. Selecting the right input data and executing individual steps in a data reduction sequence is often carried out manually by the end user. This process requires significant expertise and effort. It is also error prone and raises issues of reproducibility and record keeping. Automating data reduction workflows is therefore highly desirable for individual science reduction efforts, and mandatory for mass production.
A challenge for automating data reduction is that different data reduction goals require different specific processing workflows. For example, for quality control a processing workflow might entail the processing of all calibration data of a specific type. If the goal is instead to process science data, only a subset of the calibrations may be needed. The processing workflow in this case should select only the calibration data that are best suited for the processing of the science, process them, and then use them in combination with the science data to create the final data product. This processing of science data might be done in different ways, depending on their final usage. For example, a processing workflow might skip some processing steps in order to obtain higher signal-to-noise ratios at the expense of an increased incidence of artefacts. Another example is that distortion correction by resampling of imaging data are necessary when images are used for astrometry, but not desired when the same images are used to search for weak lensing signals. Different processing workflows are also required for unsupervised processing of science data for a science archive and interactive processing for a specific purpose. The latter case often involves executing a recipe repeatedly, with different values for parameters, to optimise the results before continuing with the next workflow steps.
To address these different data processing needs, ESO has long maintained several different workflow systems. For science users, ESO Reflex (Freudling et al. 2013) is offered as an interactive environment. For each supported pipeline a pre-packaged data processing workflow is provided that users can customise and execute on their own data. ESO Reflex includes a host of features for data organisation, data visualisation, and bookkeeping. In addition to ESO Reflex, ESO uses several other data reduction systems that run workflows for different quality control levels and for archive processing. These systems are only used within the ESO computing environment and cannot easily be exported. ESO’s different systems all run the same pipeline recipes, but each is highly tuned towards running a specific kind of workflow.
2.2 Unification of data processing workflows
It is clear that different workflows do not necessarily require different systems to run them. A sufficiently flexible system could execute different workflows that explicitly encode the different data processing scenarios as outlined above. However, such explicit specifications would largely be variants of the same underlying processing cascade. While there are substantial differences between the different workflows, their design is constrained by the inter-dependencies of the recipes on each other. For example, if a recipe is designed to process some flat-fields using preprocessed bias data, this implies a fixed processing sequence. The bias data have to be processed before the flat recipe is executed. This has to be taken into account in all workflows. In that sense, processing workflows for the different use cases differ, but are also highly repetitive in their design. As a consequence, a change in the underlying pipeline could require changing all workflows in a very similar manner and a substantial effort would be needed to keep the different processing workflows in sync. The fragility and huge effort in coding and maintaining our huge collection of workflows motivated us to look for a more robust and sustainable system for creating workflows.
The goal of this paper is to present a system where all the information necessary to execute recipes is specified only once, and workflows for different processing scenarios can then automatically be derived based on the available data, configuration files, and user input. Such a system exploits the fact that each reduction recipe requires specific data types as input, and produces specific data types as output. The recipes depend on each other in ways that are defined by their input and output. These dependences are therefore identical for all data processing. For example, a flat-field recipe will always require raw flat-field frames and a combined bias frame as input, and will produce a combined flat-field frame as output.
The basic idea of the proposed system is to specify the dependences of the recipes that are in common to any possible workflow written for a given set of recipes using a formal domain specific language. Individual processing workflows are then derived by specifying a ‘target recipe’ to process a pool of data. The target recipe is the recipe that produces the intended final output product of a processing request. For example, if the target recipe is the flat-fielding recipe, the system will derive the necessary processing sequence that in this simple example consists of running first the bias processing recipes followed by the flat-fielding recipe. By contrast, if the target is just the bias recipe, only that recipe is executed.
Figure 1 illustrates the concept of a target for the case of a somewhat complex data reduction workflow, which is a simplified version of the workflow for the X-shooter instrument on ESO’s Very Large Telescope (VLT), discussed in more detail in Sect. 4.6. The recipes are shown as ellipses, and the output of recipes that is fed to subsequent recipes are shown as arrows. All but the two recipes called spec_combination and spec_combination_std also need raw data as input which is not shown in the figure.
The recipe that takes a raw spectral image of the science object and produces a one-dimensional extracted spectrum is called science. A subsequent recipe called spec_combination combines multiple extracted spectra from the same target. In addition, there are a number of calibration recipes that include the usual bias, dark, and flat recipes that combine individual frames to produce calibration files to be applied to the data by subsequent recipes. Other calibration recipes are the fit_order recipe that finds the location of spectral orders, the wavelength_cal recipe that derives the wavelength solution from an arc lamp image, and the flexure recipe that computes the correction for instrument flexure from a special calibration with pin holes.
A noteworthy feature of the workflow is the processing of the standard star. The response recipe uses the raw data from a flux standard star, and produces a response curve to be applied to the science spectrum. In standard science processing, the spectrum of the standard star is only used for that purpose. However, in some cases the spectrum of the star is also of scientific interest, and in those cases the spectrum should be fed to the science recipe. The response curve is then computed from a different observation of a standard star. The science recipe is therefore placed twice on the workflow specification, once called science and once called science_std. Each is followed by a separate instance of the recipe to combine spectra. In that manner, each instance of the science recipe or the combination recipe can be specified as a target separately. The different instances refer to the same recipes, but are fed with different input data.
This set-up can be used to nicely illustrate the use of the target concept. In standard science processing, the desired outcome is the combined spectra of science observations. In this case, the target is the spec_combination recipe that follows the science recipe. This is illustrated in the left tree in Fig. 1. The standard star is only used to create the response curve. The right tree in the figures shows the case where the purpose of the processing is to produce a science spectrum of the standard star. Since standard stars are typically bright, a combination of the individual extracted spectra is not always desired. In that case the science_std recipe is chosen as the target. Recipes that are executed in each of the two scenarios can automatically be determined, and are shown in green in the figure.
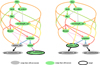 |
Fig. 1 Illustration of how the target determines the processing workflow. The two trees represent the same workflow specification when used with different targets. The ellipses represent recipes, and the arrows represent the output from one recipe that is fed to subsequent recipes. The target recipe in each case is indicated by the black outline. The ellipses for recipes that are executed are in green; non-executed recipes are in grey. The corresponding file association step is shown in Fig. 2 (see text for a detailed explanation). |
3 Data classification and organisation
A data processing workflow by itself is not sufficient to carry out a full data reduction. In order to do so, the correct data need to be identified and fed to the individual recipes. The processing itself is therefore preceded by a step that selects the data to be processed, and passes them to the right recipes. Throughout this paper, we use the term ‘data reduction workflow’ to include both the data selection and the subsequent data processing steps.
Each of the recipes of the processing workflow shown in Fig. 1 needs to be fed with a specific data type. The data type of a file can be derived from the data header based on some rules.
In the simplest case, a single header keyword or a combination of them identify the file type. The process of determining the file type of each available file is called ‘classification’.
Once all available files are classified, a subset of them can be chosen for processing. The process of selecting input files starts with the selection of input for the target recipes, and then follows the processing cascade upstream. The first step in organising the input for each recipe is to group the principal input files (i.e. the input files the recipe is designed to reduce). Hereafter, we refer to these files as the ’main input’ of a recipe. For example, the spec_combination recipe shown in Fig. 1 is designed to process together all the spectra taken of the same object. The processing of the main input requires additional input files which have to be properly selected according to some criteria. These additional files are called ‘associated files’, and the rules to select them are called ‘association rules’. An example of this would be to select a raw flat frame for the science processing based on the properties of the spectra to be processed by the science recipes. This is illustrated in Fig. 2, which shows the tree of association rules that correspond to the workflows of Fig. 1. Each folder in the figure represents a set of input files, and the lines represent the rules to select them. For example, there is a rule to select the best bias for the arc spectrum that is identified in the figure. This rule needs as input some header information from the ARC_SPECTRUM files, such as the time of observation and the binning of the detector. The rule might state that the RAW_BIAS must have the same binning as the ARC_SPECTRUM, and is as close as possible in observing time. This rule is then applied to the data to select the best match among all the provided RAW_BIASes.
It becomes clear that the description of data organisation is similar to the workflow itself, with the flow of information in the opposite direction. In simple cases, the data reduction workflow can therefore be derived from the description of the data organisation. However, as discussed in detail in Freudling et al. (2013) and shown in Sect. 4.6, in general this is not possible. In that work, we opted therefore for an independent description of the workflow and the data organisation and this approach was implemented in the ESO Reflex software. There are two disadvantages in this approach. First of all, the two descriptions are at least a partial duplication of information. In addition, while the descriptions are different, they have to be consistent with each other. Implementing and maintaining this consistency requires significant manual effort.
In this paper, we adopt a more efficient approach by first specifying the complete data processing cascade, and then adding the data organisation information, without any duplication, directly to the data processing tree from which objective-specific workflows can be derived. A specific implementation of this is discussed in Sect. 4. Such a design maintains the full flexibility to design complicated workflows, as discussed in Freudling et al. (2013), while at the same time avoiding any duplication in the description of the whole system. The consistency between the processing cascade and data organisation is inherent in the design. The description of the data processing cascade and data organisation rules can be used to derive complete data reduction workflows (i.e. the specific sequence of data selection and data processing steps necessary to process a given set of data and a specified target).
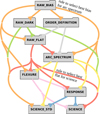 |
Fig. 2 Association rules for the workflows shown in Fig. 1. Each folder symbol represents a set of raw data. Each line represents a rule for selecting the raw files at the end of the line. The arrow at the start of the line indicates where the information to select the files is taken from. The labels on two of the lines spell out the nature of those rules. |
4 EDPS workflows
A general workflow system that implements the concepts discussed above consists essentially of three parts. A description of the processing cascade including the data organisation, an engine that parses this description, derives the appropriate workflow and applies the rules to a given set of input data to determine a sequence of recipe runs, and an executor that schedules and executes the recipes.
Such a system, compatible with ESO’s data reduction pipelines, has been implemented as the ESO Data Reduction System (EDPS). EDPS is entirely implemented in the Python language (Van Rossum & Drake 2009), including the specification of processing cascades and the accompanying data organisation rules. EDPS allows the specification of processing cascades in a clear and concise manner that describes the data reduction process without the need for detailed knowledge of the implementation of other parts of EDPS or the Python language. In this section we describe the specification of processing cascades and data organisation rules in a level of details that illustrates the basic principles. For a full description of all feature on a more technical level we refer to Zampieri et al. (2024).
4.1 Tasks
The central aspect of a processing cascade is the specification of the recipes to be executed. This is typically the first step in writing a processing cascade because the recipes and their inputs and outputs are mostly known already at the design stage of a pipeline. In EDPS the execution of a recipe is specified in ‘tasks’. Tasks are descriptions of processing steps that can be executed independently once all their inputs are available. Most tasks are used to execute recipes, but the concept of a task is more general and allows any Python code to be executed as part of the data reduction workflow. The most basic properties of a task to be specified are the recipe to be executed and its input. As noted in Sect. 3, each recipe has a main input and possibly one or more associated inputs that are needed to process the main input. A fully functionally example code-snippet of a task definition in EDPS is the following:
1 flat_task = (task(’FLAT’) 2 .with_recipe(’flat_recipe’) 3 .with_main_input(raw_flat) 4 .with_associated_input(bias_task) 5 .build())
Each task has a name for presentation purposes, in this example it is called ‘FLAT’. At execution time, the target task of the execution can be specified referring to the task by this name. Tasks have many optional parameters, some of which are fairly complex to instantiate. Creating those objects using Python constructors would require a long list of arguments. To improve readability we use the builder pattern syntax to provide values only for selected arguments. These arguments can be recognised by their starting pattern .with_. The call of the .build() method indicates that the configuration is complete.
4.2 Data sources
The input of a task as shown in the above listing is either the output from a preceding task or some files that are available at the start of the execution of a workflow. The former can be referred to by their object names, which is ‘flat_task’ in the example above. The latter have to be Flexible Image Transport System (FITS) files (Wells et al. 1981) that can be classified by header keywords. In the processing cascade, these files have to be defined as a ‘data source’, that includes a ‘classification’ rule that specifies how to recognise a file. In its simplest form it specifies one or several keywords, and their values to be used to define a data type. An example of such a simple classification rule is the following:
1 type_flat = classification_rule(’rawflat’,
2 {’filetyp’: ’RAW_FLAT’})
This classification rule specifies that file is reported as ‘rawflat’ to the recipe if the header keyword ‘filetyp’ has the value ‘RAW_FLAT’. In general, much more complex classification rules can be specified, including support for alternatives and conditional statements to analyse the headers of the files. Data from complex instruments often require more complicated rules such as these. The classification rules can then be used to define data sources, e.g.:
1 raw_flat = (data_source(’RAW_FLAT’) 2 .with_classification_rule(type_flat) 3 .build())
The above code-snippet specifies a data source named ‘RAW_FLAT’, and the corresponding files are recognised by the classification rule given above. A full specification of data sources would also include a rule for how the files should be grouped (i.e. how the collections of RAW_FLATs that are fed to each execution of a recipe are assembled). EDPS also provides a syntax to specify existing files as an alternative to the input created by a preceding task. This feature allows running the same workflow using pre-processed input files, for example using processed calibration files that are available in the ESO Science Archive Facility1 as an option when downloading the data.
4.3 Processing cascades
In most cases, designing the basic processing cascade is the first task for the development of a pipeline. The EDPS syntax allows the user to specify and visualise this overall architecture of a pipeline leaving out details such as the data classification and selection. This can be achieved by specifying the tasks with their inputs and the corresponding data sources, using the syntax described in the previous sections. An example of a processing cascade with three tasks is shown in the following listing:
1 # - data sources - 2 3 raw_bias = (data_source(’RAW_BIAS’) 4 .build()) 5 6 raw_flat = (data_source(’RAW_FLAT’) 7 .build()) 8 9 raw_science = (data_source(’RAW_SCIENCE’) 10 .build()) 11 12 # - tasks - 13 14 bias_task = (task(’bias’) 15 .with_main_input(raw_bias) 16 .build()) 17 18 flat_task = (task ( ’ flat ’) 19 .with_main_input(raw_flat) 20 .with_associated_input(bias_task) 21 .build()) 22 23 science_task = (task(’science’) 24 .with_main_input(raw_science) 25 .with_associated_input(bias_task) 26 .with_associated_input(flat_task) 27 .build())
A visualisation of this processing cascade, which can be created by EDPS, is shown in Fig. 3. Further details can be added later, turning the design into a fully functional code for the execution of a multitude of different workflows. The simplicity and readability of the specification of the basic processing cascade is an appealing property of the EDPS syntax.
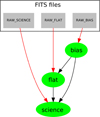 |
Fig. 3 EDPS visualisation of a simple workflow corresponding to the code listing in Sect. 4.3. The green ovals are the tasks, grey squares the data sources, red arrows the main inputs and black arrows the associated inputs of the tasks. |
4.4 Data organisation
As explained in Sect. 3, a full data reduction workflow needs additional specifications to allow the selection of associated input that corresponds to the main input of each task. This selection is, in most cases, based on the closeness in time of the associated and main input of a task, but usually also includes other conditions, such as a match in detector binning or other data properties. The way the data are associated is in most cases a property of the associated input, but in some cases different rules need to be applied depending on the task that receives the input. Association rules in EDPS can therefore either be attached to the data source or to the associated input of a task. In the simplest case the syntax of these rules is similar to that of the classification rules shown above. These associated calibrations are in some cases optional; in other words, using them might improve the quality of a reduction, but it is not strictly necessary. In other cases the calibrations are needed only under certain conditions. Such cases can be expressed in the rules and are taken into account in the data organisation.
The design of data organisation rules is an important consideration in the design of a processing cascade, with a potentially large impact on the overall quality of the data reduction. Taking optimal calibration data with astronomical instruments is often expensive in terms of observing time. A trade-off between obtaining more calibrations or investing more time in the observations of the science object is often unavoidable. This then has implications for the selection rules to be applied. An example of such a situation is that calibration data ideally are from the same night. If an instrument is sufficiently stable, then data from the previous or following nights up to some limit can be used for the calibration. Another example is that a calibration product can be replaced with a different inferior data product. The EDPS data organisation rules can capture these situations by attaching several association rules to a data source or task. Each of the rules can be assigned a quality code, and the achieved quality of the data selection is reported at run time for each dataset.
4.5 Advanced features
The features of the EDPS syntax described in the previous Sects. 4.1–4.4 are an implementation of the general concepts described in Sect. 2.1. A wide variety of data reduction processing cascades and workflows can be described with them, allowing EDPS to support the pipelines of all of ESO’s operational instruments, as well as many now-retired instruments. EDPS also supports a number of advanced features that can be used to implement more complex scenarios.
All the data organisation rules based on FITS header keywords discussed in the previous section can also be formulated directly as Python functions, bypassing the provided custom classes. This allows the implementation of complex logic that acts on combinations of keywords. This feature is useful, for example, if a pipeline supports multiple instruments or instrument modes, each of them creating data with different header structures.
Another scenario that frequently occurs is that recipe parameters have to be set differently for input data with different properties. An example of this is that if a dedicated sky observation is available, the parameters for estimating the sky levels are set differently than in the case where the sky level has to be derived from the science exposure itself. A related scenario is that the processing cascade itself depends on properties of input data (i.e. different tasks are executed for data with different properties), an example being a recipe to combine data is only triggered if a minimum number of exposures are available. EDPS supports such data-driven processing cascades. Also in this case, complex logical functions can be used in the specification of the parameters. A related feature is to allow the specification of conditions for data sources to be used.
EDPS processing cascades can also have parameters that provide the user with choices on the data reduction workflow. One use case of workflow parameters is to set parameters for several recipes simultaneously in cases when these recipe parameters need to be set consistently. Another use case is to provide users with choices in the reduction flow (see Sect. 4.6 for an example).
One supported feature that does not add any functionality, but improves the readability of EDPS processing cascades is the possibility to group tasks into subworkflows. A subworkflow is itself a processing cascade with several tasks, and can be used within the main processing cascade similar to a task. A sub-workflow can also include other subworkflows in a hierarchical manner.
4.6 Example X-shooter processing cascade
To illustrate the combination of concepts discussed in the previous sections, we show in Fig. 4 how they are used for the pipeline of the complex X-shooter instrument (Vernet et al. 2011). The topology and basic steps of this processing cascade are identical to the ones shown in Fig. 1, but this figure uses a representation of a processing cascade that shows addition information for each task and subworkflow. EDPS can create such a graph alternatively to the previously shown graph in Fig. 3. Each box represents the task or subworkflow named in the coloured header for that box.
The X-shooter instrument has three different arms, and can be used in three different observing modes, namely ‘stare’, ‘nod’, and ‘offset’. While the overall data flow for the reduction of data taken with the different modes is similar, they use different recipes for many of the steps. Some of the steps are only used for one or two of the arms. In total, the data reduction for the X-Shooter instrument uses 18 recipes that take 65 different types of files as input. Despite this complexity, using subworkflows, the main processing cascade can be displayed in a compact manner that is easy to understand by grouping recipes that do similar steps for different modes into subworkflows.
Each task and subworkflow box in Fig. 4 lists the recipe and its main inputs for each step. A task includes only one recipe, whereas a subworkflow might include several tasks and therefore several recipes. For each task and subworkflow, the different associated inputs are listed, which correspond to the coloured lines that go into that step. Finally, static calibrations are listed. Static calibrations are files such as line lists that are not tied to a specific observation and may not even be directly acquired at the telescope or by the instrument. For each associated input and static calibration, it is shown whether they are mandatory input (‘Mand’) and whether they are conditional (‘Cond’) (see Sect. 4.2). Many of the input files in the X-Shooter processing cascade are conditional because they are only used for some of the arms. For example, the near-IR arm does not require bias or dark frames as input.
This processing cascade is also an example of a case where the data reduction workflow cannot be derived from the description of the data organisation alone. The reason for this is that in some cases the optimal flat for the standard star is the one used by the science frame so that any flat-fielding inaccuracies cancel out (see Freudling et al. 2013). The general rule of ‘use a flat as close as possible in time to the file to process’ therefore does not apply in that case. This is implemented as the subworkflow flat_strategy that does not include any regular recipe, but only conditional copy commands. A workflow parameter controls which strategy is used to associate the flat frames, either the conventional ‘close in time’ rule or the strategy just described.
5 Execution of workflows
5.1 Software architecture
The EDPS processing cascades as discussed in the previous section can be coded in Python without a detailed knowledge of the inner workings of EDPS. The core functionalities of EDPS are to apply the data organisation rules to the available data, derive the data processing workflow based on the processing cascade description and the specified target, and schedule the execution of recipes.
The EDPS software has been implemented with a client-server architecture (Zampieri et al. 2024). The server part is a persistent process that carries out all the necessary processing based on the processing cascade specification, the data organisation rules and the input data. The server program does not terminate after the processing is completed, but keeps listening for requests to process data. The advantage of this approach is that the start-up processes carried out by the server need to be executed only once, and data that might be used in subsequent processing are kept in memory. This improves the overall responsiveness of EDPS after the initial start-up.
The server acts on requests that are submitted via a REST API as used by standard web service clients. These requests can in principle be submitted via a web browser, but an EDPS client is provided as a more convenient interface. Requests include the location of the data, the processing cascade specification, at least one target, and the value of any recipe or workflow parameters that should be set to a value different from the default. After receiving a request, individual processing jobs are created and executed by the EDPS server. For a future release, we anticipate the implementation of a dedicated graphical user interface (GUI) for more convenient interactive processing.
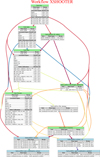 |
Fig. 4 EDPS visualisation of the full X-shooter processing cascade specification. The topology of the processing cascade is essentially the same as in Fig. 1, with one extra task called ‘flat_strategy’. Each step in Fig. 1 is now expanded with the full information. The individual steps in Fig. 1 are either a single task or a subworkflow with several tasks, as indicated in the header of each box. The colour-coding of the header distinguishes between calibration steps that produce calibration products (green) and science steps that could serve as science target (blue). The subworkflow flat_strategy does not produce any products itself and only serves to redirect products produced by other steps. |
5.2 Processing jobs
The complete processing cascade description discussed in the previous section allows EDPS to process a given set of data with the specified pipeline. The input data pool is typically a directory on a local disk or a list of files.
The kind of processing carried out on these files crucially depends on the specified target in a processing request. We want to reiterate the importance of the central concept of the target. If the target of the workflow shown in Fig. 3 is the science task, all files that match the rules for the data source RAW_SCIENCE will be processed. Files of type RAW_FLAT will only be processed if they are needed to process the selected RAW_SCIENCE files. Files of type RAW_BIAS are processed if they are needed either for the processing of the RAW_FLATs or the RAW_SCIENCEs. By contrast, if for example the bias task is specified as target, all files of type RAW_BIAS are processed irrespective of whether they are needed by any other task.
When the EDPS request is submitted by the client, the server classifies all the input files and organises them according to the encoded rules in order to enable the processing of the main input of the target task. All the information needed to execute each individual data processing step is collected in a ‘job’. Jobs are individual processing units that can be executed once all input data for that job are available. The information contained in a job includes the recipe to execute; the main and any associated inputs; the recipe parameters; and general attributes, such as a task name and the completeness of the input files. A single task can generate multiple jobs from a request, depending on how many groups of files need to be processed. For example, if a request includes two science files and associated calibration files and specifies the science task of the processing cascade depicted in Fig. 3 as the target, then the following jobs will be generated:
- 1.
Two jobs from the science task, one for each of the RAW_SCIENCE files to be processed;
- 2.
One job from the flat task for each group of RAW_FLATS files to be processed. If the two RAW_SCIENCE files require two different sets of RAW_FLATS, then two jobs are generated;
- 3.
One job from the bias task for each group of RAW_BIAS to be processed. Depending on whether the biases for the different RAW_FLATS and RAW_SCIENCE tasks are the same or not, the bias task could thus generate up to four jobs.
If all the mandatory input data for a job are available or can be produced by the execution of other jobs, a job is marked as ‘complete’. If this condition is not met, a job is ‘incomplete’. If a job is incomplete, the job and all of its child jobs cannot be executed (see Sect. 5.3).
5.3 Job execution and scheduling
The jobs created by EDPS then need to be scheduled for execution. An obvious constraint on the scheduling is that a job cannot be executed before all of its input are available. If the input is the output of other jobs, they have to be scheduled first. EDPS will also detect whether a recipe has been executed with the same set of parameters and input files before, and in such a case re-use the output produced by the previous execution. This is commonly referred to as ‘smart re-runs’.
These constraints by themselves do not imply a unique sequence of scheduling. Hardware resources and the general objectives of the scheduling have to be considered in order to choose a strategy for scheduling jobs. If a large amount of data is to be processed in batch mode, the objective usually is to minimise the total processing time, given a set of hardware resources. However, the situation is different in interactive mode where a user wants the jobs to be executed in a sequence that allows interaction in a manner that they can easily follow and understand the consequences of their interactions. However, the situation is different in interactive mode where user input is required after some steps. In that situation, the request for user input should be presented in an order that is easy to follow and in a way that the consequences of the interactions can easily be understood. A typical usage scenario for the interactive case is that the goal is to process several independent sets of files with the target recipe. An example of this is to process several sets of images. Each set includes images that overlap, but the individual sets are spatially separated from each other. An image co-adding recipe accepts one full set of overlapping images. Following Freudling et al. (2013), EDPS uses the concept of datasets, which are the files that are processed together by the target recipes, together with all calibration files that are needed to make the execution of the target recipe possible. An interactive user will then want to process one dataset at a time, inspecting the intermediate data products as the processing proceeds. The processing is paused after the execution of each recipe to allow the review of the products and possible re-execution of this step with different recipe parameters. During such an interactive processing, jobs that process data for other datasets should only be executed if they do not slow down the processing of the currently considered dataset. Typically, processing one dataset at a time will increase the overall processing time for all datasets. The benefit of this sequence is the shortened waiting time for the user to provide the interactive input for the first dataset.
EDPS supports three algorithms to define the order of processing before the start of any job execution, and one that schedules the jobs dynamically. The static algorithms are the ‘depth-first’ (DFS), ‘breadth-first’ (BFS), and ‘type search’ algorithms. The DFS scheduling covers the interactive processing use case described above. The BFS schedules jobs independent of datasets or types, taking into account the number of jobs that can be simultaneously submitted for execution. The type search scheduling schedules all jobs of a certain input type in sequence. The em dynamic scheduling means that the order is determined after the start of job executions. When a job is completed, all jobs in the waiting queue are considered for execution. Depending on the total number of cores and CPUs made available to the server and the number of threads used by each recipe, jobs that make the best use of the available resources are started.
5.4 Testing of processing cascades
EDPS processing cascades with many tasks, data sources, and associated data organisation are sufficiently complex to warrant extensive testing when changes are made. EDPS processing cascades are therefore accompanied by a specification of data files that can be used to test the processing cascades. The corresponding FITS files are then created when the test is executed. The test itself consists in using the data organisation of the processing cascade to produce the input for and trigger the tasks, without actually executing the recipes themselves. The tests therefore verify the grouping and association of data as well as the structure of the processing cascades, thereby identifying the most common errors in processing cascades. The test specifications are coded in JavaScript Object Notation (JSON) files that describe the input files with their header keywords, the target tasks, and the values of any processing cascade parameters. In addition, the test specifications include the expected jobs and their input files. The EDPS test facility runs the processing cascade specification on the automatically created test files and compares the list of tasks that have been triggered and their inputs with the expected outcome as specified in the test itself. A test is passed if all predicted tasks are triggered with their predicted inputs and no unexpected tasks are triggered. These tests can, for example, be automatically executed in an integrated development environment (IDE), and they can also be run automatically by a continuous integration system any time a change to a processing cascade is made to check for any regression.
6 Summary
We propose a data reduction system that automatically creates data reduction workflows from a processing cascade specifications. The necessary specifications can be written following the typical development cycle of a data reduction pipeline, where the reduction steps are specified first and the data organisation information is added later as needed.
The system avoids duplication of the specifications in two different ways: 1) all information to derive workflows for different reduction use cases is encoded in a single description (called the processing cascade) in an efficient way; 2) the specification of the data organisation is fully integrated into the processing cascade definition. This is different from previous systems, where the processing workflow follows the data organisation, which has to be specified first, and is therefore limited in its flexibility, or the approach advocated in Freudling et al. (2013) where the data organisation is specified completely independent of the processing workflow.
EDPS is an implementation of such a system. It was designed to run ESO’s data reduction pipelines, but workflows can be written for any pipeline that follows the basic principle of stand-alone recipes that are executed in sequence, use FITS files for input and output, and can be configured using parameter files. EDPS has been in routine operation at ESO since April 2023, and was publicly released in October 20232. This release web page includes code, documentation, and installation instructions.
Acknowledgements
The EDPS team at ESO includes Lodovico Coccato, Wolfram Freudling, Enrique Garcia, Andrea Modigliani, Ahmed Mubashir Khan, Ralf Palsa, Stanislaw Podgorski, Martino Romaniello, and Stefano Zampieri.
References
- Brodheim, M. N., O’Meara, J. M., Mader, J. A., et al. 2022, SPIE Conf. Ser., 12186, 121860H [NASA ADS] [Google Scholar]
- Freudling, W., Romaniello, M., Bramich, D. M., et al. 2013, A & A, 559, A96 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Lacy, M. 2016, SPIE Conf. Ser., 9910, 991007 [Google Scholar]
- Lacy, M., An, R., Benson, J., et al. 2015, in ASP Conf. Ser., 495, Astronomical Data Analysis Software and Systems XXIV (ADASS XXIV), eds. A. R. Taylor, & E. Rosolowsky, 425 [Google Scholar]
- Law, D. R., Cherinka, B., Yan, R., et al. 2016, AJ, 152, 83 [Google Scholar]
- Lemoine-Busserolle, M., Comeau, N., Kielty, C., Klemmer, K., & Schwamb, M. E. 2019, AJ, 158, 153 [NASA ADS] [CrossRef] [Google Scholar]
- Lockhart, K. E., Do, T., Larkin, J. E., et al. 2019, AJ, 157, 75 [NASA ADS] [CrossRef] [Google Scholar]
- Romaniello, M., Arnaboldi, M., Barbieri, M., et al. 2023, The Messenger, 191, 29 [NASA ADS] [Google Scholar]
- Tran, H. D., Burger, M., & Hack, W. 2020, in ASP Conf. Ser., 527, Astronomical Data Analysis Software and Systems XXIX, eds. R. Pizzo, E. R. Deul, J. D. Mol, J. de Plaa, & H. Verkouter, 587 [Google Scholar]
- Van Rossum, G., & Drake, F. L. 2009, Python 3 Reference Manual (Scotts Valley, CA: CreateSpace) [Google Scholar]
- Vernet, J., Dekker, H., D’Odorico, S., et al. 2011, A & A, 536, A105 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Weilbacher, P. M., Palsa, R., Streicher, O., et al. 2020, A & A, 641, A28 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Wells, D. C., Greisen, E. W., & Harten, R. H. 1981, A & AS, 44, 363 [NASA ADS] [Google Scholar]
- Zampieri, S., et al. 2024, EDPS Manual (Garching: ESO) [Google Scholar]
All Figures
 |
Fig. 1 Illustration of how the target determines the processing workflow. The two trees represent the same workflow specification when used with different targets. The ellipses represent recipes, and the arrows represent the output from one recipe that is fed to subsequent recipes. The target recipe in each case is indicated by the black outline. The ellipses for recipes that are executed are in green; non-executed recipes are in grey. The corresponding file association step is shown in Fig. 2 (see text for a detailed explanation). |
| In the text | |
 |
Fig. 2 Association rules for the workflows shown in Fig. 1. Each folder symbol represents a set of raw data. Each line represents a rule for selecting the raw files at the end of the line. The arrow at the start of the line indicates where the information to select the files is taken from. The labels on two of the lines spell out the nature of those rules. |
| In the text | |
 |
Fig. 3 EDPS visualisation of a simple workflow corresponding to the code listing in Sect. 4.3. The green ovals are the tasks, grey squares the data sources, red arrows the main inputs and black arrows the associated inputs of the tasks. |
| In the text | |
 |
Fig. 4 EDPS visualisation of the full X-shooter processing cascade specification. The topology of the processing cascade is essentially the same as in Fig. 1, with one extra task called ‘flat_strategy’. Each step in Fig. 1 is now expanded with the full information. The individual steps in Fig. 1 are either a single task or a subworkflow with several tasks, as indicated in the header of each box. The colour-coding of the header distinguishes between calibration steps that produce calibration products (green) and science steps that could serve as science target (blue). The subworkflow flat_strategy does not produce any products itself and only serves to redirect products produced by other steps. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


