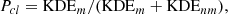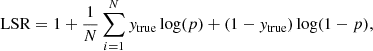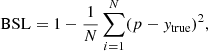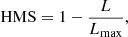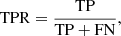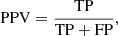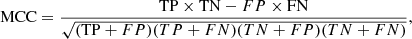| Issue |
A&A
Volume 650, June 2021
|
|
|---|---|---|
| Article Number | A109 | |
| Number of page(s) | 13 | |
| Section | Numerical methods and codes | |
| DOI | https://doi.org/10.1051/0004-6361/202040252 | |
| Published online | 15 June 2021 | |
pyUPMASK: an improved unsupervised clustering algorithm
1
Instituto de Astrofísica de La Plata (IALP-CONICET), 1900 La Plata, Argentina
e-mail: msolpera@gmail.com
2
CENTRA, Faculdade de Ciências, Universidade de Lisboa, Ed. C8, Campo Grande, 1749-016 Lisboa, Portugal
3
Facultad de Ciencias Exactas, Ingeniería y Agrimensura (UNR), 2000 Rosario, Argentina
4
Instituto de Física de Rosario (CONICET-UNR), 2000 Rosario, Argentina
5
Facultad de Ciencias Astronámicas y Geofísicas (UNLP-IALP-CONICET), 1900 La Plata, Argentina
Received:
28
December
2020
Accepted:
24
March
2021
Aims. We present pyUPMASK, an unsupervised clustering method for stellar clusters that builds upon the original UPMASK package. The general approach of this method makes it plausible to be applied to analyses that deal with binary classes of any kind as long as the fundamental hypotheses are met. The code is written entirely in Python and is made available through a public repository.
Methods. The core of the algorithm follows the method developed in UPMASK but introduces several key enhancements. These enhancements not only make pyUPMASK more general, they also improve its performance considerably.
Results. We thoroughly tested the performance of pyUPMASK on 600 synthetic clusters affected by varying degrees of contamination by field stars. To assess the performance, we employed six different statistical metrics that measure the accuracy of probabilistic classification.
Conclusions. Our results show that pyUPMASK is better performant than UPMASK for every statistical performance metric, while still managing to be many times faster.
Key words: open clusters and associations: general / methods: data analysis / open clusters and associations: individual: NGC 2516 / methods: statistical
© ESO 2021
1. Introduction
Galactic open clusters are of great importance for the study of the Galaxy’s chemical evolution, structure, and dynamics; these sources also provide test beds for astrophysical codes that model the evolution of stars. Located largely on the disk of the Milky Way, analyses of open clusters is severely hindered by the presence of contaminating field stars, located in the foreground and background of the object of interest. These stars are projected on the observed field of view and end up deeply mixed with the cluster members. The process of disentangling these two classes of elements, of members from nonmembers (i.e., field stars), can be referred to as “decontamination”.
A proper decontamination of the cluster region is a key previous step to the analysis of the cluster sequence in search of fundamental parameters (e.g., metallicity, age, distance and extinction) that characterize the open cluster. This analysis, which is often performed in photometric space, requires a sequence that is as complete as possible, but also as free of contaminating field stars (nonmembers) as possible. The goal of a decontamination algorithm is to obtain a subset of stars that fulfills both these conditions simultaneously.
Over the years, a handful of decontamination algorithms have been presented in the stellar cluster literature. Most of these are variations of the Vasilevskis-Sanders method (Vasilevskis et al. 1958; Sanders 1971) applied over proper motions, which are generally considered to be much better member discriminators than photometry. Nonparametric approaches have also been developed (Cabrera-Cano & Alfaro 1990; Javakhishvili et al. 2006) and even an interactive tool to determine membership probabilities was presented (Balaguer-Núñez et al. 2020)1. More references regarding membership estimation methods can be found in Krone-Martins & Moitinho (2014, henceforth KMM14) and Perren et al. (2015).
The Unsupervised Photometric Membership Assignment in Stellar Clusters algorithm (UPMASK), originally presented in KMM14, has the advantage of being not only nonparametric, but also unsupervised. This means that no a priori selection of field stars is required to serve as a comparison model, which is generally the case in the previously mentioned methods. Although UPMASK was motivated by the need of assigning cluster memberships from photometric data, KMM14 had pointed out that the method is general and could be easily applied to other data types and clusters of objects. Recent examples of UPMASK used on proper motions (and parallax data) can be found in Cantat-Gaudin et al. (2018a, b, 2019), Carrera et al. (2019), and Yontan et al. (2019).
In the six years since its publication, the KMM14 article has been referenced almost 50 times; this work has also been applied to stellar proper motions and to study clusters of galaxies, which indicates a wide adoption by the astrophysical community.
In this work we present an improved version of the original UPMASK algorithm, which we call pyUPMASK because it is written entirely in Python. We believe this new package can be of great use, particularly with the advent of the recent early data release 3 (eDR3, Gaia Collaboration 2021) of the Gaia mission (Gaia Collaboration 2016). This package is made available as a stand-alone code, but it will also be included in an upcoming release of our Automated Stellar Cluster Analysis tool (ASteCA, Perren et al. 2015).
Throughout the article we refer to statistical clusters as simply clusters and explicitly distinguish them from stellar clusters when required.
This paper is organized as follows: In Sect. 2 we give a brief summary of the UPMASK algorithm and present the details of the enhancements introduced in our code. Section 3 introduces the synthetic cluster sample used in the analysis, and describes the selected statistical performance metrics employed to assess the behavior of UPMASK and pyUPMASK. The results are summarized in Sect. 4. Finally, our conclusions are given in Sect. 5.
2. Methods
We present a brief description of the general algorithm used in UPMASK as well as the major enhancements introduced in pyUPMASK. Both methods are open source and their codes can be found in their respective public repositories23.
2.1. The UPMASK algorithm
The UPMASK package is described in full in KMM14 and we do not repeat it in this work. We give instead a summary of the most relevant parts and of its core algorithm. The original article provides a more detailed description.
Assigning probability memberships to the two classes of elements within a stellar cluster field (members and field stars) is a notably complicated problem for two main reasons. First, the classes are usually very much imbalanced. This means that one of the classes (field stars) can make up a lot more than 50% of the total dataset. In some extreme cases, the frame of an observed stellar cluster can consist of over 90% of field stars and less than 10% of actual true members. Even worse, this information (i.e., the true balance) cannot be assumed to be known a priori. Second, the two classes are deeply entangled. This is particularly true in the two-dimensional coordinates space where members and field stars are mixed throughout the entire cluster region. Off the shelf clustering methods normally assume that there is some kind of frontier that largely separates the classes with minimal overlap. This is not the case in stellar clusters analysis.
The UPMASK algorithm deals with both of these issues in a clever and effective way, by taking advantage of the fact that we can approximate the distribution of field stars in the coordinates space with a uniform model. This is further discussed in Sect. 2.2.2.
The UPMASK algorithm is composed of two main blocks: an outer loop and an inner loop. The outer loop is responsible for taking into account the uncertainties in the data and rerunning the inner loop a manually fixed number of times; these uncertainties are optional and turned off by default. The latter is required because of the inherent stochasticity of the K-means (KMS) method (MacQueen 1967), employed by the inner loop. The number of runs for the outer loop is one of the two most important parameters in the algorithm.
The inner loop holds the two main parts that make up the core of the algorithm: the clustering method (KMS, as stated before), and the random field rejection method (henceforth: RFR). The clustering method is applied on the nonpositional features (e.g. photometry and proper motions), and separates the cluster data into N clusters. The N value is determined by a parameter that determines the number of elements that should be contained in each cluster. That is, dividing the total number of stars by this value gives N, the final number of clusters that are generated.
After the clustering method is applied, the RFR method serves the purpose of filtering those clusters identified by the KMS that are consistent with a random uniform distribution of elements. This consistency is assessed in UPMASK by means of a two-dimensional kernel density estimation (KDE) analysis. In short: the KDE of the coordinates space of each cluster (identified by the KMS in the previous step) is compared with the KDE of a two-dimensional uniform distribution in the same range. If these are deemed to be similar enough, the cluster is discarded as a realization of a random selection of field stars, and all its stars are assigned a value of 0. Those clusters that survive the RFR process are kept for a subsequent iteration of the inner loop. When no more clusters are rejected and the inner loop is finished, all the stars within surviving clusters are assigned a value of 1. After this, a new iteration of the outer loop is initiated.
The final probabilities assigned to each star are simply the averages of the (0, 1) values assigned by the inner loop at each run of the outer loop.
The two parameters mentioned above are the most important parameters in UPMASK, since varying their value can substantially affect the performance of the method. We comment on how we selected these parameters in Sect. 3.3.
2.2. The pyUPMASK algorithm
An obvious difference between pyUPMASK and UPMASK is that the former is written entirely in Python4 instead of R5, as is the case with UPMASK. We believe that this is a considerable advantage given the noticeable shift of the astrophysical community toward the Python language in recent years. This is made evident by large Python-based projects such as Astropy6 (Astropy Collaboration 2013, 2018) and international conferences such as Python in Astronomy7. A recent survey found that Python is the most popular programming language in the astronomical community (Momcheva & Tollerud 2015; Tollerud et al. 2019).
The general structure of pyUPMASK closely follows the UPMASK algorithm: an outer loop containing an inner loop that applies the cluster identification and rejection methods. What sets these two algorithms apart is twofold: First, pyUPMASK supports almost a dozen clustering methods, while UPMASK only supports KMS; and second, pyUPMASK contains three added analysis blocks that are not present in UPMASK. In Fig. 1 we show the complete flow chart of the pyUPMASK algorithm. The blocks indicated in violet are those that are either enhanced or added in this work.
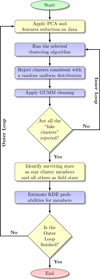 |
Fig. 1. Flow chart of the pyUPMASK code. The enhanced clustering block and the analysis blocks added in this work are indicated in violet. |
The enhanced clustering methods block and the three added blocks are detailed in Sects. 2.2.1–2.2.4, respectively. The remaining portions of the code are mostly equivalent to those described in KMM14 for UPMASK and, for the sake of brevity, we do not repeat their details or purpose in this work.
2.2.1. Clustering methods
While UPMASK supports the KMS method exclusively (as of the current version 1.2), pyUPMASK relies on the Python library scikit-learn8 (Pedregosa et al. 2011) for the implementation of most of the supported clusterings methods. This library includes around a dozen different clustering methods for unlabeled data, which are all available to use in pyUPMASK. Eventually this can be extended to support even more methods in future releases of the code via the PyClustering library9.
Once chosen, the clustering method processes the nonspatial data at the beginning of the inner loop as shown in Fig. 1. The number of individual clusters to generate is fixed indirectly through a user-selected input parameter, as done in UPMASK. Each of these clusters is then analyzed by the RFR method and kept or rejected given its similarity with a random uniform distribution of elements. This is further discussed in Sect. 2.2.2.
In Sect. 4 we present a suit of tests performed with four of the methods provided by scikit-learn: KMS, mini batch k-means (MBK, Sculley 2010), Gaussian mixture models (GMM, Baxter et al. 2010), and agglomerative clustering (AGG, Zepeda-Mendoza & Resendis-Antonio 2013). In addition to these we include tests performed with two methods developed in this work: the nearest neighbors density method (KNN), which is based on the density peak approach introduced in Rodriguez & Laio (2014); and the Voronoi (VOR) method, which is based on the construction of N-dimensional Voronoi diagrams (Voronoi 1908). The latter three methods (AGG, KNN and VOR) have a characteristic in common: no stochastic process or approximation is employed by any of them. In other words, these methods are deterministic. This means that, for the same input data and input parameters, different runs lead to one single result. Assuming that no data resampling is performed (the default setting in both UPMASK and pyUPMASK) the outer loop then needs to be run only once because subsequent runs would produce the same probabilities each time. For this reason we refer to these as “single-run” methods. As can easily be inferred, these are significantly faster than UPMASK and the rest of the tested methods, which require multiple outer loop runs.
The results obtained with the six selected methods are compared to UPMASK results obtained on the same dataset of synthetic clusters. The synthetic clusters dataset is described in Sect. 3.1.
2.2.2. Ripley’s K function
After the clusters are generated on the nonspatial data, the RFR block is used to filter out those that are consistent with the realization of a random uniform distribution on the spatial data (i.e., coordinates). The hypothesis at work is that field stars are randomly scattered throughout the two spatial dimensions of the frame, following somewhat closely a uniform distribution. Actual star cluster members, on the other hand, present a more densely packed spatial distribution. The latter is of course an approximation to the real, and unknown, probability distribution of field stars, but it is still a very reasonable one as the results show.
The UPMASK algorithm employs a KDE-based method to characterize the distribution of each cluster found in the spatial dimensions. This distribution is then compared to that of thousands of random uniform distributions generated in the same two-dimensional range and with the same number of elements. After that, a “KDE distance” is obtained by comparing their means, maximum, and standard deviation values. If the distance between both distributions is less than a user-defined threshold parameter, the cluster is considered to be close enough to a realization of a random uniform distribution. When this condition is met, the cluster is rejected as a “fake cluster” (see Fig. 1).
In pyUPMASK we introduce Ripley’s K function (Ripley 1976, 1979) to assess the closeness of a cluster to a random uniform distribution. This function is defined as
where A is the area of the domain (our observed frame), N is the number of points within it, dij is the distance between points i, j, I is a function that results in 1 if the condition is met and 0 otherwise, eij is the edge correction (if required), and r is the scale at which the 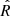 function is calculated.
function is calculated.
Ripley’s K function is employed to test for complete spatial randomness (CSR), also called homogeneous Poisson point process, which basically consist of points randomly located on a given domain. In a two-dimensional space it is trivial to prove that if points are distributed following CSR, then K(r) equals πr2 (Streib & Davis 2011). The K function is thus a perfect match for our intended usage which is precisely to test if a set of points (stars) are distributed following uniform spatial randomness. We employ the form of the K function given by
which converges to r under CSR. Following Dixon (2014) we combine information from several distances (r values) in a single test statistic defined as
where sup is the supremum. Given that the lengths of the observed frame are normalized by default to the range [0, 1] prior to processing, the list of distances at which Eq. (3) is calculated are chosen to be in the range [0, 0.25]. This is the range advised in the Kest function of the spatstat package (Baddeley et al. 2015)10.
The null hypothesis (H0) for the 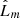 is that the points follow CSR. We need to select a critical value such that if the test is greater than that value, the test is considered to be statistically significant and H0 is rejected. Such critical values were estimated by Monte Carlo simulations in Ripley (1979). The pyUPMASK algorithm uses the 1% critical value; that is, there is a 1% probability of erroneously rejecting H0 (also called a Type I error). This critical value is approximated for
is that the points follow CSR. We need to select a critical value such that if the test is greater than that value, the test is considered to be statistically significant and H0 is rejected. Such critical values were estimated by Monte Carlo simulations in Ripley (1979). The pyUPMASK algorithm uses the 1% critical value; that is, there is a 1% probability of erroneously rejecting H0 (also called a Type I error). This critical value is approximated for 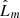 as
as 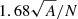 , where A and N are the area and number of points, respectively. In future releases of the code we plan on integrating analytical expressions for the critical values, for example, those obtained in Lagache et al. (2013) and Marcon et al. (2013).
, where A and N are the area and number of points, respectively. In future releases of the code we plan on integrating analytical expressions for the critical values, for example, those obtained in Lagache et al. (2013) and Marcon et al. (2013).
The pyUPMASK algorithm employs the astropy implementation of the K function, which includes the required edge corrections for points that are located close to the domain boundaries. Compared to the UPMASK KDE test, the K function is not only a more natural choice for this task, it is also orders of magnitude faster.
2.2.3. Gaussian-uniform mixture model
After the RFR block is finished and the fake clusters are rejected, only those stars that were found in clusters sufficiently different from a random uniform distribution of points are kept. This dataset of stars is nonetheless still affected by contamination from field stars that could not be removed. This is because these field stars were, by chance, associated with a cluster composed mainly of true star cluster members and thus not rejected. We developed a method to clean this region, applied to the two-dimensional coordinates space that we call GUMM, because it is based on fitting a Gaussian-uniform mixture model to the dataset. This can be thought of as a simpler version of the spatial plus proper motions space modelization found in previous works, for example, Jones & Walker (1988).
A D-dimensional Gaussian distribution can be written as
where x is the D-dimensional data vector, and (μ,Σ) are the mean and covariance matrix. A GMM with K components (i.e., Gaussians) is defined as
where πi are the weights (or mixing coefficients) associated with each of the K components. Similar to the GMM, we define the GUMM as a two-dimensional mixture model composed of a Gaussian, representing the stellar cluster, and a uniform distribution, representing the noise due to contaminating field stars. The full model is then written as
where U[0, 1] is the uniform distribution in the range [0, 1], and πx(x = 0, 1) are the unknown weights for each model. No restrictions are imposed on the position, shape, or extension of the 2D Gaussian representing the stellar cluster. Following the recipe employed by the classic GMM, we use the iterative expectation-maximization algorithm (EM, Dempster et al. 1977) to estimate these weights as well as the mean and covariance of the 2D Gaussian. After the EM algorithm converges to a solution, each star is assigned a probability of belonging to the 2D Gaussian (i.e., to the putative cluster). We then need to decide which stars to reject as field stars based on these probability values. To do this the percentile distribution of the probabilities is generated and the value at which the curve begins a sharp climb toward large probabilities is automatically identified as the probability cut. The value corresponding to the climb in the percentile curve is estimated with the method developed in the kneebow package11. The user can input a manual value for this probability cut (or even skip the GUMM altogether), but after extensive testing this method has proven to give very good results and it is thus the recommended default. Stars below this value are rejected as contaminating field stars and the surviving stars are kept as cluster members.
The results of processing a group of stars from a synthetic cluster with the GUMM can be seen in Fig. 2. The plot in panel a shows the 2D coordinates space after the RFR block rejects those clusters consistent with a random uniform distribution. It can be seen that, even after clusters mainly composed of field stars are rejected, the central overdensity is still visibly contaminated by the surrounding field stars. The plot in panel b shows the probabilities assigned to each star of belonging to the 2D Gaussian via the GUMM process. In the plot in panel c, we show the percentile diagram for the probabilities, where the red line shows the value at which the cut is imposed. Finally, the plot in panel d shows the region after those stars with probabilities below the aforementioned cut are rejected.
 |
Fig. 2. GUMM process in four steps. Panel a: the set of stars that survived the RFR block. Panel b: probabilities assigned by the GUMM to all the stars in the frame. Panel c: the method for selecting the probability cut value using a percentile plot. Panel d: the final set cleaned from most of the contaminating field stars. |
This process, although almost trivial at first glance, greatly improves the purity of the final sample of estimated true members at very little cost regarding completeness. The hypothesis at work is of course that the putative stellar cluster is more concentrated in the coordinates space than regular field stars, as previously stated.
2.2.4. Kernel density estimator probabilities
Once a run of the inner loop is finished, each star in the observed field is classified to be either a cluster member or a field star. Although continuous (spatial) probabilities are assigned in the GUMM step, these are used to apply a coarse classification between members and nonmembers. The information that moves on to the next segment is the hard binary classification. This means that only probability values of 0 and 1 are assigned up to this stage. The KDE block takes these binary probabilities and turns them into continuous probabilities in the range [0, 1]. This improves the final results in general by assigning more realistic probability values. Furthermore, this block is essential for single-run clustering methods (defined in Sect. 2.2.1). Clustering methods such as KMS or GMM require multiple outer loop runs. The final probabilities are then estimated by averaging all the binary probability values, which breaks the binarity. Single-run methods work, as the name indicates, on a single run of the outer loop. This means that without this block, single-run methods would assign probabilities of 0 and 1 exclusively.
The KDE probabilities are assigned after a full run of the inner loop, with all stars classified as either members or nonmembers. The process is as follows:
-
Separate each of those two classes into different sets.
-
Estimate the KDE for each class using all the available data, that is, coordinates plus the data dimensions used for clustering (photometry, proper motions, etc.).
-
Evaluate all the data in the frame in the KDE obtained for each class.
-
Use the above evaluations as likelihood estimates in the Bayesian probability for two exclusive and exhaustive hypotheses (i.e., a star belongs to either the members distribution or the field stars distribution).
The final cluster membership probability (using uniform equal priors) is written as
where KDEm and KDEnm are the KDE likelihoods for the members and nonmembers (field), respectively. The process can be seen in Fig. 3 for the coordinates dimensions (even though it is applied on all the data dimensions, described in Sect. 3.1). The plot in panel a shows the two classes, members and nonmembers, generated after the inner loop is finished. In the plot in panel b, we show the two-dimensional coordinates KDEs for both classes, noting again that this is applied on all the data dimensions. The plot in panel c shows the nonbinary Pcl probabilities assigned by the method in the coordinates space. Finally, the plot in panel d is equivalent to the plot in panel c, but for the proper motions space.
 |
Fig. 3. KDE probabilities method shown in the coordinates space. Panela: members and nonmembers, as estimated by the inner loop process. Panel b: KDEs for both classes. Panel c: final Pcl probabilities assigned in the coordinates space. Panel d: same as panel c, but for proper motions. |
3. Validation of the method
In order to perform a thorough comparison of the performance of pyUPMASK with that of UPMASK, we applied both methods to a large number of synthetic clusters and quantified the results using numerous statistical metrics. In this section, we describe the set of synthetic clusters, the selected metrics, and the reasoning behind the choice of input parameters.
3.1. Synthetic datasets
We employed a total of 600 synthetic clusters to analyze the performance of UPMASK and pyUPMASK, the latter in the six configurations mentioned in Sect. 2.2.1. This set is divided into a subset of 320 clusters, and another of 280 clusters. The first subset is equivalent to that used in the original UPMASK article (KMM14) in the sense that it is composed of clusters with synthetic photometry generated with the same process as that used in KMM14. We refer to this subset as PHOT hereinafter. The second subset contains 280 clusters generated by adding synthetic proper motions to all the stars in the frame; we refer to this subset as PM hereinafter. The idea is to see how the two algorithms handle the case in which only photometry is available (i.e., the PHOT dataset), and the increasingly common case (thanks to the Gaia mission) in which proper motions with very reasonable quality are available (i.e., the PM dataset). The performance of UPMASK and pyUPMASK is tested using the 600 synthetic clusters obtained by combining the PHOT and PM datasets.
Clusters were generated with a wide range of field star contamination. The level of contamination is measured by the contamination index (CI), which is defined as the number of field stars to cluster members in the frame to match the “contamination rate” used in KMM14. The maximum CI in our set of synthetic clusters is 200.
In Fig. 4 we show examples of a PHOT (top) and PM (bottom) synthetic clusters, which are generated with moderate contamination (CI ≈ 50).
 |
Fig. 4. Top row: coordinates and CMD for a PHOT synthetic cluster with moderate CI. Bottom row: coordinates and vector-point diagram for a PM synthetic cluster with moderate CI. |
3.2. Performance metrics
A proper choice for evaluating the classification performance of a probabilistic model (such as UPMASK or pyUPMASK) is a debate that carries on even today (Hand 2009; Hernández-Orallo et al. 2012). Different metrics or scoring rules yield different results regarding the performance of the model (Merkle & Steyvers 2013), which means that relying on a single metric is not recommended. This is particularly true when dealing with datasets that can be highly imbalanced, as is our case. We thus chose to employ multiple metrics. By combining all of these, we expect to obtain a non-biased assessment of the overall performance of pyUPMASK versus UPMASK.
We selected six metrics that can be divided into two groups of three each. The first group consists of strictly proper scoring rules, which guarantee that they are only optimized when the true classification is obtained. This group is composed of the following metrics:
Logarithmic scoring rule:
where N is the number of elements, ytrue ∈ {0, 1} is the true label, and p = Pr(y = 1) is the probability that y = 1, that is, the probability that the element belongs to the class identified with a 1 (Good 1952). The LSR (also called log-loss or cross-entropy) heavily penalizes large differences between ytrue and p.
Brier score loss:
which is equivalent to the mean squared error for binary classification; it was originally introduced in Brier (1950).
H measure:
where L is the loss function, and Lmax is the maximum loss; the expression for the loss function is much too mathematically involved to be presented here, it can be seen in full in Hand (2009). This is a relatively new metric. It was developed as a replacement of the popular AUC (area under the receiver operating characteristic curve) score; now known to be an incoherent performance measure and thus not recommended (Lobo et al. 2008; Parker 2011; Hand & Anagnostopoulos 2014). The HMS automatically handles unbalanced classes by treating the misclassification of the smaller class (in our case almost always true members, except for extremely low CI values) as more serious than those of the larger class.
It is worth noting that the definitions of LSR and BSL were altered from their original forms by multiplying by -1 and adding plus 1. This way all the metrics defined assign 1 to a perfect score.
The three metrics in the first group can be used directly on the membership probabilities in the [0, 1] range, resulting from UPMASK or pyUPMASK.
The second group defined below consists of scoring rules that are applied to binary classifiers. These are the types of metrics used in the original KMM14 article and we employ them in this work for consistency12. In the definitions that follow TP is a true positive (a member star correctly classified as such), TN is a true negative (a field star correctly classified as such), FN is a false negative (a member star incorrectly classified as field), and FP is a false positive (a field star incorrectly classified as member):
True positive rate:
which is also called sensitivity or recall; it measures the proportion of true members that are correctly identified.
Positive predictive value:
which is also called precision; it measures how many stars classified as members are true members.
Matthews correlation coefficient:
which was introduced in Matthews (1975); it can be thought of as an equivalent to Pearson’s correlation coefficient for binary classifiers. Unlike the TPR and PPV, the MCC also takes the TNs into account. It is recommended when dealing with imbalanced classes, as is our case.
To turn the problem into one of binary classification and to be able to use the three metrics defined in the second group, we must first select a probability threshold that separates the stars into the members and nonmembers classes. In KMM14 a single threshold of 90% was used. Since the choice of a threshold can affect the results from these three metrics, we decided to use the following two different thresholds: 50% and 90%. This way we end up with the following nine metrics to test the performance of UPMASK and pyUPMASK: LSR, BSL, HMS, TPR5, PPV5, MCC5, TPR9, PPV9, and MCC9; where the subindex 5 and 9 indicate the 50% and 90% thresholds, respectively.
3.3. Input parameters selection
There are two main parameters in UPMASK and pyUPMASK that affect the outcome of the methods: the number of stars per cluster and the number of runs of the outer loop. The former, which we refer to as Nclust, was investigated in KMM14, in which the authors concluded that a value between 10 to 25 is appropriate. In the latest version (v1.2) of the UPMASK code, depending on how it is run, the default value for Nclust is either 25 or 5013.
We performed our own tests using 100 synthetic clusters (50 PHOT and 50 PM) covering the full CI range, selected at random from the full list of 600 mentioned in Sect. 3.1. This set was analyzed with the nine performance metrics described in Sect. 3.2. In Fig. 5 we show the results obtained for three Nclust values 15, 25, and 50. We combined all the metrics for the 100 synthetic clusters into one set and subtracted these (900) values for a given Nclust value from another. The results are plotted versus the CI of the synthetic clusters. From panels a to c the combinations N15 − N25, N15 − N50, and N25 − N50 are shown, where a positive value means that the Nclust value on the left performed better than the value on the right, and vice versa for negative values. As can be seen, the differences are rather small and do not tend to change for different CI values. We thus decided to use the middle value Nclust = 25 for all the UPMASK and pyUPMASK runs, as a reasonable number of default stars per cluster for all the CI range.
 |
Fig. 5. Panel a–c: boxplot of the combined metrics difference vs. CI for the 100 synthetic clusters used in the test. Combinations for the N15, N25, N50 values are shown. Panel d: outer loop convergence analysis. The convergence percentile of the nine metrics vs. the number of outer loop run is shown. The black dashed line indicates the 90% convergence point. |
Deciding how many times the outer loop should run is the other important parameter: a low number terminates the code before it is able to present fully converged probability values and a large number wastes processing time. We processed the same set of 100 synthetic clusters with Nclust = 25 and analyzed when each of the nine metrics converged to a stable value. The stabilization point is defined as the outer loop run where the metric changes inside the ±0.025 range for five consecutive runs. The results are shown in Fig. 5d. plot as a the convergence percentile (i.e., the percentage of clusters that have converged) for each metric versus the outer loop run. Almost all the metrics reach a convergence above 90% before the 25th outer loop run. The two exceptions are TPR9 and PPV9, which still show a convergence above 85% before the 25th run. Given these results we use 25 runs in the outer loop for all the UPMASK and pyUPMASK analyses with the obvious exceptions of the single-run methods described in Sect. 2.2.1.
The PHOT set was processed using all the available photometry as input (V, B − V, U − B, V − I, J − H, H − K) but selecting only the four principal dimensions after the principal component analysis dimensionality reduction. For the PM set we used the proper motions (μα, μδ), with no dimensionality reduction. Proper motions are generally regarded as better cluster members discriminators than photometry, owing to the rounded shape of its distribution in contrast with the irregular shape of the sequence of a cluster in the photometric space.
4. Results
To ensure that the results are comparable between the pyUPMASK and UPMASK runs, all the analyses were performed on the same computer cluster. In what follows, the results are classified according to whether pyUPMASK or UPMASK performed better for a given metric and synthetic cluster. We allow for a small range of ±0.005 to act as a “tie zone” in which the two methods can be thought of as performing equally well. In Appendix A we show the results of comparing pyUPMASK with the Bayesian method included in AsteCA. These are not included here because the methods are not directly comparable, as explained in the Appendix.
In Figs. 6 and 7 we show the metrics for the 320 and 280 synthetic clusters in the PHOT and PM datasets, respectively, for each of the six clustering methods used in pyUPMASK. The blue, yellow, and red bars depict the proportion of cases for which pyUPMASK performed better, equally well, and for which UPMASK performed better, respectively. It is easy to see that, although with some variation across clustering methods, pyUPMASK has a better performance than UPMASK for all the methods and all the metrics, particularly for the PM dataset. This is an outstanding result that unmistakably shows the large improvement brought by pyUPMASK. The three methods that apply multiple outer loop runs (MBK, KMS, GMM) show a clear advantage over the remaining single-run methods, regarding the proportion of cases for which pyUPMASK resulted in larger metric values.
 |
Fig. 6. Results for the 320 synthetic clusters in the PHOT dataset processed with the six clustering methods used in pyUPMASK vs. UPMASK. For each metric, the blue and red bars represent the cases where pyUPMASK and UPMASK performed better, respectively. The yellow bars represent cases in which both methods performed equally well. |
In Cantat-Gaudin et al. (2018a) the authors used a modified version of UPMASK to estimate membership probabilities for more than 1200 cataloged clusters. The modification was motivated by the need to increase the speed for processing large numbers of clusters. This modification mainly consists in replacing the default KDE based method in the RFR block in UPMASK for a minimum spanning tree method (see article for more details). We tested this modified version14, which we refer to as MST, using the same set of synthetic clusters and metrics employed so far. The code was executed with 25 runs of the outer loop and 15 stars per cluster; internal tests showed that this gave more adequate results than using 25.
The results of our six clustering methods, plus the MST method, versus UPMASK can be compressed into a single matrix plot as shown in Fig. 8. We show the X minus UPMASK percentage metric difference, where X represents each of the pyUPMASK clustering methods plus MST. This value is obtained subtracting the number of synthetic clusters, where pyUPMASK/MST performed better than UPMASK, from the number of clusters where UPMASK showed a better performance, and taking the percentage. This difference ranges from -100, which would indicate that UPMASK performed better on all 600 synthetic clusters, to 100, indicating that pyUPMASK (or MST) was the better performer for the 600 clusters. A value of 0 indicates that both methods performed better on an equal number of cases.
 |
Fig. 8. Matrix plot of the six pyUPMASK clustering methods plus MST, vs. the nine defined metrics for the 600 synthetic clusters. Each square shows the percentage difference of the number of cases for which pyUPMASK/MST performed better, minus the number for which UPMASK performed better. |
As can be seen, for the pyUPMASK methods all the squares in the matrix are positive (the smallest being the PPV5 metric for the VOR method), which again shows that pyUPMASK performed significantly better than UPMASK, measured by any of the employed metrics. The advantage of the MBK, KMS and GMM methods over the single-run methods is easier to see here compared to Figs. 6 and 7. The only exception is the TPR9 metric for which the VOR, KNN, and AGG methods show a larger differential than the remaining multiple-runs methods; that is, more true members are classified as such. This comes at the expense of the PPV9 metric, for which the MBK, KMS and GMM methods show much larger values; that is, fewer field stars are incorrectly classified as members. Other than this, there is no visible relation between any clustering method and a given performance metric.
The MST method shows a somewhat erratic behavior across the metrics. It performs worse than UPMASK for almost all of the clusters for several metrics (i.e., HMS, TPR5, MCC9 and TPR9) and better for a few others (e.g. LSR and BSL). Overall, the statistical performance of the MST method is worse than UPMASK and pyUPMASK with any of the tested clustering methods. Notwithstanding, MST is faster than UPMASK (as we show below) and outperforms all other methods in the LSR and BSL metrics.
In Fig. 9 we show the dependence with CI for the pyUPMASK minus UPMASK difference for all the metrics, for each clustering method. A positive value (green region) means that pyUPMASK performed better, while a negative value (red region) means that it performed worse than UPMASK. The PHOT and PM sets are shown with triangles and circles, respectively. There is no apparent trend with CI for the results of any clustering method. What is clear is that pyUPMASK performs even better for the PM set as evidenced by the overall larger (more positive) differences, particularly for clusters with large CI values. This is a very desirable result taking into account that high quality proper motions are becoming more accessible very fast.
 |
Fig. 9. Differences between pyUPMASK vs. UPMASK results for all the metrics combined, vs. the CI (shown as a logarithm). Each clustering method is shown separately, as are the PHOT and PM sets using blue triangles and black circles, respectively. The red and green regions correspond to the regions for which pyUPMASK gives worse and better results than UPMASK, respectively. |
We can further compress the results by combining each metric into a single value, for each of the clustering methods tested in pyUPMASK. That is, we take the 5400 results for each clustering method (600 synthetic clusters times nine metrics) and calculate the percentage at which pyUPMASK outperformed UPMASK. The same process can be applied to the synthetic clusters for which UPMASK outperformed pyUPMASK to obtain a similar, inverted, percentage. The results are shown in Table 1. This table shows that even the worst pyUPMASK performer (VOR) gives better metrics than UPMASK 66% of the times. The method with the highest pyUPMASK percentage (GMM) outperforms UPMASK 83% of the times, which is a massive advantage. The worst individual metric result is obtained for TPR9 in the MBK method. Still the value is larger than 50%, which means that the majority of the cases were better handled by pyUPMASK. On the other end of the analysis the best metric result is found for HMS in the GMM method, for which pyUPMASK manages to outperform UPMASK for virtually all of the cases.
Aggregated results for all the metrics and all the synthetic clusters, for the six pyUPMASK clustering methods used, as percentage of results where pyUPMASK outperformed UPMASK, and vice versa, respectively.
Another important aspect along with the performance measured by the statistical metrics is the performance measured in computing time. This is shown in Fig. 10 as a bar plot normalized to the total time used by UPMASK to process the 600 synthetic clusters. The numbers on top of the bars display how many times faster each clustering method in pyUPMASK is compared to UPMASK. We also show the time performance of the MST modification mentioned previously. The fastest method is expectedly a single-run method, KNN, which performs 170 times faster than UPMASK. This is an enormous margin of difference. Even the slowest method, GMM, is faster than UPMASK: this method manages to process the set of synthetic cluster employing almost 38% less time than UPMASK or 1.6 times faster. On average we can say that pyUPMASK using a single-run method is over 100 times faster than UPMASK and is more than 3 times faster for the multiple run methods.
 |
Fig. 10. Amount of time employed in processing the 600 synthetic clusters by each pyUPMASK method (blue bars), the MST method (orange bar), and UPMASK (red bar). The bars are normalized so that UPMASK corresponds to a total value of 1. |
The choice between which clustering method to employ in pyUPMASK then depends on the specific requirements of the analysis. If the absolute best performance measured by a classification metric is sought after, then clearly GMM is the method to chose (with the advantage of being faster than UPMASK). If we can trade some performance for a faster process, then KMS or MBK can be used. And if we are willing to trade even more classification performance, while still performing much better than UPMASK, then VOR, KNN, or AGG are by far the fastest approaches.
Finally, we consider the issue of computational resources requirements. We found that for very large input data files memory and processing power requirements can be too much for most methods to handle. Although the VOR clustering method is the worst performer out of the six tested methods (measured by classification metrics), it has an advantage compared to all the others, including UPMASK, when it comes to analyzing large files.
To obtain the Voronoi diagram of an N-dimensional set of points, the Python scipy package relies on the Qhull library (Barber et al. 1996)15. This library is written in the C language which makes it extremely efficient, thus making the pyUPMASK VOR method very efficient for large datasets.
To test this we downloaded a large 6 × 6 deg region around the NGC2516 cluster from the Gaia second data release (Gaia Collaboration 2018). The resulting field contains over 420 000 stars up to a maximum magnitude of G = 19 mag. This limit was imposed because beyond this value the photometric errors grow exponentially.
The frame was processed with the six tested pyUPMASK clustering methods and UPMASK, using proper motions and parallax as input data. We used 25 outer loop iterations for all the methods, except of course for the single-run methods, and a value of 25 for the parameter that determines the number of elements per cluster (i.e., the default values for both parameters as explained in Sect. 3.3).
Only three methods were able to complete the process: VOR, KNN, and MBK. The methods AGG, KMS, and GMM failed owing to memory requirements as they attempted to allocate arrays of ∼640 Gb, ∼31 Gb, and ∼31 Gb on memory, respectively. The UPMASK algorithm was not able to finish even the first iteration of the inner loop within the first iteration of the outer loop after a full week of running, so it was halted. The results of the VOR, KNN, and MBK methods can be seen in a color-magnitude diagram (CMD) in Fig. 11. We plotted the 1500 stars with larger membership probabilities given by each method, as this is the approximate number of cluster members in the frame (given by a simple stellar density analysis). It is evident that the VOR method returns the most reasonable and less contaminated CMD out of the three. Furthermore, this method managed to process the cluster almost 4 and 40 times faster than KNN and MBK, respectively. It is worth noting that on a personal computer, which has far less resources than a computational cluster, VOR was the only method that was able to run.
 |
Fig. 11. Results for the NGC2516 cluster by the VOR (left), KNN (center), and MBK (right) methods. Estimated members are shown as green circles; the field stars are shown as gray dots. |
A smaller field containing this same cluster was analyzed with UPMASK in Cantat-Gaudin et al. (2018a). The processed area contains only ∼1100 stars associated with this cluster up to a magnitude of G = 18 mag. The analysis done in this work resulted in less than 800 stars with membership probabilities (MPs) above 0.5 and ∼100 stars with MPs > 0.9. In contrast, using the same magnitude cut, we are able to obtain with the VOR method on our very large field over 1700 stars with MPs > 0.99 tracing a well-defined sequence. The advantage of studying a cluster using almost all of its members versus using less than 10% of the members (comparing the large MPs subsets), is obvious.
The VOR method is thus the only one that was able to produce quality results for this very large dataset, and it did so while using the least amount of processing time by a wide margin.
5. Conclusions
Since its development in KMM14 the UPMASK code has been used to analyze thousands of clusters. This is because it is a very smart, general, and efficient unsupervised method, that requires no prior knowledge about the observed field.
In this work we introduced pyUPMASK, a tool based on the general UPMASK algorithm with several added enhancements. The primary aim of pyUPMASK is the assignment of membership probabilities to cluster stars observed in a frame contaminated by field stars.
We tested our code extensively using 600 synthetic clusters affected by a large range of contamination. Six performance metrics were employed, three of which were in two different configurations, to ensure sufficient coverage when assessing statistical classification. The results from six different clustering methods in pyUPMASK were compared to those from UPMASK. Under the conditions established for the analysis, the pyUPMASK tool proved to clearly outperform UPMASK while still managing to be faster (and, for the single-run methods, extremely faster).
This new tool is thus highly configurable (around a dozen clustering algorithms supported), fast, and an excellent performer measured by several metrics. The pyUPMASK algorithm is fully written in Python and is made available for its use under a GPL v3 general public license16.
Clusterix 2.0: http://clusterix.cab.inta-csic.es/clusterix/
pyUPMASK: https://github.com/msolpera/pyUPMASK
Acknowledgments
The authors would like to thank the anonymous referee, for their helpful suggestions and corrections to the manuscript. The authors would like to thank Dr Anagnostopoulos for his help with the hmeasure (https://github.com/cran/hmeasure) R package. The authors would like to thank Dr Cantat-Gaudin for sharing the code used in Cantat-Gaudin et al. (2018a). M.S.P., G.I.P., and R.A.V. acknowledge the financial support from CONICET (PIP317) and the UNLP (PID-G148 project). A.M. acknowledges support from the Portuguese Fundação para a Ciência e a Tecnologia (FCT) through the Strategic Programme UID/FIS/00099/2019 for CENTRA. This research has made use of NASA’s Astrophysics Data System. This research made use of the Python language (van Rossum 1995) and the following packages: NumPy (http://www.numpy.org/) (Van Der Walt et al. 2011); SciPy (http://www.scipy.org/) (Jones et al. 2001); Astropy (http://www.astropy.org/), a community-developed core Python package for Astronomy (Astropy Collaboration 2013, 2018); scikit-learn (http://scikit-learn.org/) (Pedregosa et al. 2011); matplotlib (http://matplotlib.org/) (Hunter 2007). This work has made use of data from the European Space Agency (ESA) mission Gaia (https://www.cosmos.esa.int/gaia), processed by the Gaia Data Processing and Analysis Consortium (DPAC, https://www.cosmos.esa.int/web/gaia/dpac/consortium). Funding for the DPAC has been provided by national institutions, in particular the institutions participating in the Gaia Multilateral Agreement.
References
- Astropy Collaboration (Robitaille, T. P., et al.) 2013, A&A, 558, A33 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Astropy Collaboration (Price-Whelan, A. M., et al.) 2018, AJ, 156, 123 [Google Scholar]
- Baddeley, A., Rubak, E., & Turner, R. 2015, in Spatial Point Patterns: Methodology and Applications with R, (CRC Press), Chapman& Hall/CRC Interdisciplinary Statistics [CrossRef] [Google Scholar]
- Balaguer-Núñez, L., López del Fresno, M., Solano, E., et al. 2020, MNRAS, 492, 5811 [Google Scholar]
- Barber, C. B., Dobkin, D. P., & Huhdanpaa, H. 1996, ACM Trans. Math. Software, 22, 469 [CrossRef] [MathSciNet] [Google Scholar]
- Baxter, R. A. 2010, in Mixture Model, eds. C. Sammut, & G. I. Webb (Boston, MA: Springer, US), 680 [Google Scholar]
- Brier, G. W. 1950, Mon. Weather Rev., 78, 1 [CrossRef] [Google Scholar]
- Cabrera-Cano, J., & Alfaro, E. J. 1990, A&A, 235, 94 [NASA ADS] [Google Scholar]
- Cantat-Gaudin, T., Jordi, C., Vallenari, A., et al. 2018a, A&A, 618, A93 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Cantat-Gaudin, T., Vallenari, A., Sordo, R., et al. 2018b, A&A, 615, A49 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Cantat-Gaudin, T., Jordi, C., Wright, N. J., et al. 2019, A&A, 626, A17 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Carrera, R., Pasquato, M., Vallenari, A., et al. 2019, A&A, 627, A119 [CrossRef] [EDP Sciences] [Google Scholar]
- Dempster, A. P., Laird, N. M., & Rubin, D. B. 1977, J. R. Stat. Soc.: Ser. B (Methodol.), 39, 1 [Google Scholar]
- Dixon, P. M. 2014, Ripley’s K Function (American Cancer Society) [Google Scholar]
- Gaia Collaboration (Prusti, T., et al.) 2016, A&A, 595, A1 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Gaia Collaboration (Brown, A. G. A., et al.) 2018, A&A, 616, A1 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Gaia Collaboration (Brown, A. G. A., et al.) 2021, A&A, 649, A1 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Good, I. J. 1952, J. R. Stat. Soc.: Ser. B (Methodol.), 14, 107 [Google Scholar]
- Hand, D. J. 2009, Mach. Learn., 77, 103 [CrossRef] [Google Scholar]
- Hand, D., & Anagnostopoulos, C. 2014, Pattern Recognit. Lett., 40, 41 [CrossRef] [Google Scholar]
- Hernández-Orallo, J., Flach, P., & Ferri, C. 2012, J. Mach. Learn. Res., 13, 2813 [Google Scholar]
- Hunter, J. D., et al. 2007, Comput. Sci. Eng., 9, 90 [NASA ADS] [CrossRef] [Google Scholar]
- Javakhishvili, G., Kukhianidze, V., Todua, M., & Inasaridze, R. 2006, A&A, 447, 915 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Jones, B. F., & Walker, M. F. 1988, AJ, 95, 1755 [NASA ADS] [CrossRef] [Google Scholar]
- Jones, E., Oliphant, T., Peterson, P., et al. 2001, SciPy: Open Source Scientific Tools for Python, [Online; accessed 2016-06-21] [Google Scholar]
- Krone-Martins, A., & Moitinho, A. 2014, A&A, 561, A57 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Lagache, T., Lang, G., Sauvonnet, N., & Olivo-Marin, J.-C. 2013, PLoS ONE, 8 [Google Scholar]
- Lobo, J. M., Jiménez-Valverde, A., & Real, R. 2008, Global Ecol. Biogeogr., 17, 145 [CrossRef] [Google Scholar]
- MacQueen, J. 1967, Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics (Berkeley, Calif.: University of California Press), 281 [Google Scholar]
- Marcon, E., Traissac, S., & Lang, G. 2013, ISRN Ecol., 2013, 1 [CrossRef] [Google Scholar]
- Matthews, B. 1975, Biochimica et Biophysica Acta (BBA) - Protein Structure, 405, 442 [CrossRef] [Google Scholar]
- Merkle, E. C., & Steyvers, M. 2013, Decis. Anal., 10, 292 [CrossRef] [Google Scholar]
- Momcheva, I., & Tollerud, E. 2015, ArXiv e-prints [arXiv:1507.03989] [Google Scholar]
- Parker, C. 2011, 2011 IEEE 11th International Conference on Data Mining (IEEE) [Google Scholar]
- Pedregosa, F., Varoquaux, G., Gramfort, A., et al. 2011, J. Mach. Learn. Res., 12, 2825 [Google Scholar]
- Perren, G. I., Vázquez, R. A., & Piatti, A. E. 2015, A&A, 576, A6 [NASA ADS] [CrossRef] [EDP Sciences] [Google Scholar]
- Ripley, B. D. 1976, J. Appl. Probab., 13, 255 [CrossRef] [Google Scholar]
- Ripley, B. D. 1979, J. R. Stat. Soc. Ser. B (Methodol.), 41, 368 [Google Scholar]
- Rodriguez, A., & Laio, A. 2014, Science, 344, 1492 [NASA ADS] [CrossRef] [PubMed] [Google Scholar]
- Sanders, W. L. 1971, A&A, 14, 226 [NASA ADS] [Google Scholar]
- Sculley, D. 2010, Proceedings of the 19th International Conference on World Wide Web, WWW ’10 (New York, NY, USA: Association for Computing Machinery), 1177 [CrossRef] [Google Scholar]
- Streib, K., & Davis, J. W. 2011, CVPR, 2011, 2305 [Google Scholar]
- Tollerud, E. J., Smith, A. M., Price-Whelan, A., et al. 2019, Bull. Am. Astron. Soc., 51, 180 [Google Scholar]
- Van Der Walt, S., Colbert, S. C., & Varoquaux, G. 2011, Comput. Sci. Eng., 13, 22 [Google Scholar]
- van Rossum, G. 1995, Python tutorial, Report CS-R9526 [Google Scholar]
- Vasilevskis, S., Klemola, A., & Preston, G. 1958, AJ, 63, 387 [NASA ADS] [CrossRef] [Google Scholar]
- Voronoi, G. 1908, Journal für die reine und angewandte Mathematik, 1908, 97 [CrossRef] [Google Scholar]
- Yontan, T., Bilir, S., Bostancı, Z. F., et al. 2019, Ap&SS, 364 [CrossRef] [Google Scholar]
- Zepeda-Mendoza, M. L., & Resendis-Antonio, O. 2013, in Hierarchical Agglomerative Clustering, eds. W. Dubitzky, O. Wolkenhauer, K. H. Cho, & H. Yokota (New York, NY: Springer, New York), 886 [Google Scholar]
Appendix A: pyUPMASK versus ASteCA results
We present a comparison between the membership probability estimation algorithm included in ASteCA and pyUPMASK. It is worth noting that ASteCA is a complete package for processing stellar clusters that includes a Bayesian membership estimation method. This method, which has not changed since the Perren et al. (2015) article was published, is based on comparing the distributions of field stars and stars within the cluster region in whatever data space the user decides to use (photometric, proper motions, parallax, or any combination). The cluster region is defined by the center coordinates and radius values estimated by separate methods in ASteCA that were applied previous to the Bayesian membership method. The pyUPMASK method (similarly UPMASK), on the other hand, is a method for estimating membership probabilities. That is, it represents just a portion of what the ASteCA package comprises.
The reason for not including this comparison in the main article is that the Bayesian method in ASteCA and pyUPMASK are not directly comparable. Unlike UPMASK and pyUPMASK, which are unsupervised methods, the membership method included in ASteCA is supervised because it requires an a priori separation of classes. That is: the field stars, identified as those stars located in the field region, and the possible cluster members, identified as those stars located in the cluster region, must be segregated before the membership method can be applied. Hence, the membership probabilities obtained with the Bayesian method in ASteCA are a reflection not only of the method itself, but also of the separate methods used to estimate the center and radius values.
The ASteCA algorithm was thus applied on both datasets (PHOT and PM), allowing it to automatically estimate the center coordinates and radius value of the synthetic cluster. As shown in Figs. A.1 and A.2, pyUPMASK performs better than ASteCA for both datasets, particularly for the PHOT synthetic clusters. We emphasize again that these results are not directly comparable because, in the case of the ASteCA membership probabilities, we also include the performance of the center of the cluster and radius estimation methods. If any of these fail, which is not uncommon for scarcely populated clusters or those embedded in fields with large amounts of contamination, then the Bayesian membership estimation method in ASteCA fails too. This fact notwithstanding, this is another great result that demonstrates the capabilities of pyUPMASK.
All Tables
Aggregated results for all the metrics and all the synthetic clusters, for the six pyUPMASK clustering methods used, as percentage of results where pyUPMASK outperformed UPMASK, and vice versa, respectively.
All Figures
 |
Fig. 1. Flow chart of the pyUPMASK code. The enhanced clustering block and the analysis blocks added in this work are indicated in violet. |
| In the text | |
 |
Fig. 2. GUMM process in four steps. Panel a: the set of stars that survived the RFR block. Panel b: probabilities assigned by the GUMM to all the stars in the frame. Panel c: the method for selecting the probability cut value using a percentile plot. Panel d: the final set cleaned from most of the contaminating field stars. |
| In the text | |
 |
Fig. 3. KDE probabilities method shown in the coordinates space. Panela: members and nonmembers, as estimated by the inner loop process. Panel b: KDEs for both classes. Panel c: final Pcl probabilities assigned in the coordinates space. Panel d: same as panel c, but for proper motions. |
| In the text | |
 |
Fig. 4. Top row: coordinates and CMD for a PHOT synthetic cluster with moderate CI. Bottom row: coordinates and vector-point diagram for a PM synthetic cluster with moderate CI. |
| In the text | |
 |
Fig. 5. Panel a–c: boxplot of the combined metrics difference vs. CI for the 100 synthetic clusters used in the test. Combinations for the N15, N25, N50 values are shown. Panel d: outer loop convergence analysis. The convergence percentile of the nine metrics vs. the number of outer loop run is shown. The black dashed line indicates the 90% convergence point. |
| In the text | |
 |
Fig. 6. Results for the 320 synthetic clusters in the PHOT dataset processed with the six clustering methods used in pyUPMASK vs. UPMASK. For each metric, the blue and red bars represent the cases where pyUPMASK and UPMASK performed better, respectively. The yellow bars represent cases in which both methods performed equally well. |
| In the text | |
 |
Fig. 7. Same as Fig. 6 but for the 280 synthetic clusters in the PM dataset, exclusively. |
| In the text | |
 |
Fig. 8. Matrix plot of the six pyUPMASK clustering methods plus MST, vs. the nine defined metrics for the 600 synthetic clusters. Each square shows the percentage difference of the number of cases for which pyUPMASK/MST performed better, minus the number for which UPMASK performed better. |
| In the text | |
 |
Fig. 9. Differences between pyUPMASK vs. UPMASK results for all the metrics combined, vs. the CI (shown as a logarithm). Each clustering method is shown separately, as are the PHOT and PM sets using blue triangles and black circles, respectively. The red and green regions correspond to the regions for which pyUPMASK gives worse and better results than UPMASK, respectively. |
| In the text | |
 |
Fig. 10. Amount of time employed in processing the 600 synthetic clusters by each pyUPMASK method (blue bars), the MST method (orange bar), and UPMASK (red bar). The bars are normalized so that UPMASK corresponds to a total value of 1. |
| In the text | |
 |
Fig. 11. Results for the NGC2516 cluster by the VOR (left), KNN (center), and MBK (right) methods. Estimated members are shown as green circles; the field stars are shown as gray dots. |
| In the text | |
 |
Fig. A.1. Same as Fig. 6 but showing pyUPMASK versus ASteCA. |
| In the text | |
 |
Fig. A.2. Same as Fig. 7 but showing pyUPMASK versus ASteCA. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.



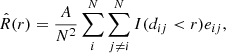
![$$ \begin{aligned} \hat{L}(r) = [\hat{K}(r)/\pi ]^2, \end{aligned} $$](/articles/aa/full_html/2021/06/aa40252-20/aa40252-20-eq3.gif)
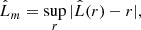
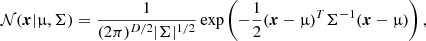
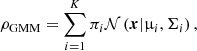
![$$ \begin{aligned} \rho _{\rm GUMM}= \pi _{0} \mathcal{N} \left(\boldsymbol{x} | \upmu , \Sigma \right)+ \pi _{1} U[0,1], \end{aligned} $$](/articles/aa/full_html/2021/06/aa40252-20/aa40252-20-eq10.gif)