A&A 392, 1129-1147 (2002)
DOI: 10.1051/0004-6361:20020760
Y. K. Ng1,2 - E. Brogt1,3 - C. Chiosi4 - G. Bertelli2,5
1 - TNO TPD Space, PO Box 155, 2600 AD Delft, The Netherlands
2 -
Padova Astronomical Observatory,
Vicolo dell'Osservatorio 5, 35122 Padua, Italy
3 -
Kapteyn Astronomical Institute, PO Box 800, 9700 AV Groningen,
The Netherlands
4 - Padova Department of Astronomy,
Vicolo dell'Osservatorio 2, 35122 Padua, Italy
5 - National Council of Research, IAS, Rome Italy
Received 14 February 2002 / Accepted 15 May 2002
Abstract
A new method, AMORE - based on a genetic algorithm optimizer,
is presented for the automated study of colour-magnitude diagrams.
The method combines several stellar population synthesis tools developed
in the last decade by or in collaboration with the Padova group.
Our method is able to recover, within the uncertainties,
the parameters
- distance, extinction, age, metallicity, index of a power-law
initial mass function and the index of an exponential star formation rate -
from a reference synthetic stellar population.
No a priori information is inserted to recover
the parameters, which is done simultaneously and not one at a time.
Examples are given to demonstrate and to better understand
biases in the results, if one of the input parameters is deliberately
set fixed to a non-optimum value.
Key words: methods: data analysis, numerical - stars: HR-diagram, statistics
The present data flow of many ongoing surveys - such as 2MASS (Beichman et al. 1998; Skrutskie 1999), DeNIS (Epchtein et al. 1997, 1999), EIS (Renzini & da Costa 1997; da Costa 1997; da Costa et al. 1998), OGLE-II (Udalski et al. 1997; Paczynski et al. 1999), SDSS (Fan 1999 and references cited therein), and even upcoming surveys as GAIA (Gilmore et al. 1998; Perryman et al. 2001) - is so large that one requires either a semi-automated or a fully automated method to analyse the colour-magnitude diagrams (CMDs) in the resulting databases. In this paper we discuss the development and the tests of an automated analysis method, which fully employs the colour and magnitude information available about the stars populating the CMD. Our method is based on an implementation of the CMD diagnostics suggested by Ng (1998). The method uses, in contrast to other techniques (see Bertelli et al. 1992; Gallart et al. 1996 & 1999; Geha et al. 1998 Harris & Zaritsky 2001; Hernandez et al. 1999; Holtzman et al. 1997 & 1999) the full, unbinned distribution of magnitudes and colours of the stars populating the CMD.
The purpose of this paper is to verify that astrophysical parameters for a synthetic single stellar population can be reliably retrieved with the so-called "AutoMatic Observation REnderer'' AMORE. In Sect. 2 an outline of AMORE is given together with its individual building blocks. In Sect. 3 we outline the method we use and in Sect. 4 we describe the tests performed with synthetic stellar populations. The results are given in Sect. 5 and we discuss in Sect. 6 the practical limits on the convergence, which is imposed by some degeneracy of the parameter space. We end with prospects on forthcoming tests, recommendations for improvements, and an outlook on future developments.
AMORE tries to find the best matching synthetic CMD to an observed CMD. Such a synthetic CMD contains for stellar aggregates the contribution of one or more stellar populations at the same distance. In the case of a CMD along a particular line of sight in our Galaxy the synthetic CMD can moreover contain the contribution of various populations with stars distributed at different distances.
In this paper we focus on the implementation and the performance of AMORE for the fitting of CMDs. For sake of argument only one, single stellar population has been considered. The implementation of automatic fitting CMDs with multiple stellar populations with stars at the same or different distances will be subject of forthcoming papers.
In the late-eighties Bertelli developed a code to generate synthetic
Hertzsprung-Russell diagrams (HRDs) from the isochrones computed by
the Padova group (cf. Chiosi et al. 1989). Initially the
synthetic colour-magnitude diagram (CMD) technique was applied mainly
in the studies of LMC open clusters![]() (see for example Bertelli et al. 1985, 1990 or Chiosi et al. 1989),
through which the amount of convective overshoot was calibrated
for the computation of a new generation of stellar evolutionary tracks.
Successive improvements were gradually applied when new sets of
evolutionary tracks (see Bertelli et al. 1994 for details)
were computed with improved radiative opacities (Iglesias et al.
1992).
(see for example Bertelli et al. 1985, 1990 or Chiosi et al. 1989),
through which the amount of convective overshoot was calibrated
for the computation of a new generation of stellar evolutionary tracks.
Successive improvements were gradually applied when new sets of
evolutionary tracks (see Bertelli et al. 1994 for details)
were computed with improved radiative opacities (Iglesias et al.
1992).
The backbone of HRD-ZVAR, the extended version of the HRD generator, is formed by the evolutionary tracks
computed by Bertelli et al. (1990;
![]() ),
Bressan et al. (1993;
),
Bressan et al. (1993;
![]() ),
and Fagotto et al. (1994a,b,c;
),
and Fagotto et al. (1994a,b,c;
![]() ,
0.004, 0.008, 0.050, 0.10).
The metallicities of the tracks follow the enrichment law
,
0.004, 0.008, 0.050, 0.10).
The metallicities of the tracks follow the enrichment law
![]() (see references cited in Chiosi
1996 and Pagel & Portinari 1998).
(see references cited in Chiosi
1996 and Pagel & Portinari 1998).
HRD-ZVAR indicates that the metallicity Z is not limited to the fixed values for which the evolutionary tracks have been computed, but is variable through interpolation between the metal-poorest and metal-richest tracks available inside the database of evolutionary tracks. In this way one is able to generate synthetic stellar populations with a smooth metallicity coverage. A prerequisite however is to use a complete and homogeneous library of evolutionary tracks and some improvements are expected if one adopts a grid of tracks with a smoother metallicity coverage.
HRD-ZVAR has been distributed (privately) to various research groups
and is also referred to as ZVAR![]() .
The version distributed, modified, and used by for example Aparicio
(1999), Gallart (1998), and Ng et al. (1996, 1997) is from now on referred to as
V1.0. Version V1.6 is used for the simulations and results presented
in this paper. This version contains a number of modifications and
improvements which speeds up the code and fixes some (rarely encountered)
bugs which interfered with the automatic minimization
process. Although various analysis methods are available, we limit
ourselves here to the description of the parameters related to the
HRD-ZVAR as adopted for AMORE.
.
The version distributed, modified, and used by for example Aparicio
(1999), Gallart (1998), and Ng et al. (1996, 1997) is from now on referred to as
V1.0. Version V1.6 is used for the simulations and results presented
in this paper. This version contains a number of modifications and
improvements which speeds up the code and fixes some (rarely encountered)
bugs which interfered with the automatic minimization
process. Although various analysis methods are available, we limit
ourselves here to the description of the parameters related to the
HRD-ZVAR as adopted for AMORE.
After selection of a set of tracks with fixed metallicity
and the choice of the parameters
![]() and
and
![]() ,
(the mass loss along the Red Giant Branch (RGB) and the Thermally
Pulsing Asymptotic Giant Branch (TP-AGB) phases respectively), the
major input to be specified for HRD-ZVAR are:
,
(the mass loss along the Red Giant Branch (RGB) and the Thermally
Pulsing Asymptotic Giant Branch (TP-AGB) phases respectively), the
major input to be specified for HRD-ZVAR are:
![\begin{figure}
\par\includegraphics[width=8.8cm,clip]{aah3492f1.eps}
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg29.gif) |
Figure 1: AMORE flowchart: schematic diagram of the individual building blocks. PIKAIA outlines the direction of the evolution paths to be investigated. Input for the stellar population synthesis engine HRD-ZVAR is the Padova library of evolutionary tracks. The luminosity and effective temperature for each synthetic star of arbitrary metallicity is then transformed to an absolute magnitude in a photometric passband with the method outlined by Bertelli et al. (1994) and Bressan et al. (1994). The synthetic HRD is then "observed'' and "detected'' through a Monte Carlo "observing run'' with the HRD-GST. Note that an alternative route is possible with single stellar populations (SSPs). The synthetic CMD is compared with the observed diagram and a fitness parameter is subsequently communicated to PIKAIA, which suggests a new set of parameters for each trial. The iteration lasts for a user defined, fixed number of trials. POWELL's method of minimization is subsequently applied to get closer to the local or global minimum. After computation of the uncertainties for each parameter the evolutionary run is either aborted or a new PIKAIA cycle is started after shrinking the limits of the parameter space (see Sect. 2.9). |
| Open with DEXTER | |
HRD-ZVAR was integrated in a galactic model by Ng (1994, 1997ab). The HRD-GST (Galactic Software Telescope) has been applied in the studies of the galactic structure towards the Galactic Centre (Ng et al. 1995, 1996 and Bertelli et al. 1995, 1996) and other regions in our Galaxy (Ng et al. 1997). In this paper we do not require the full complexity of the structural properties from the GST model. We only use a limited number of options to "observe'' a synthetic HRD at the suggested distance and to simulate the photometric errors, extinction and crowding.
A table of the photometric errors, covering a specific magnitude interval per passband, is used and the program interpolates linearly to obtain the intermediate values. We assume for the simulation that the photometric errors are Gaussian distributed. A different description of the photometric errors will be used when published artificial star tests (Stetson & Harris 1988; Gallart et al. 1999) on an observational data set are indicative for a significant deviation from a Gaussian behaviour. The visual extinction is simulated with the average value provided and appropriately scaled to a value in different passbands. In the UBVRI passbands we adopted the scaling according to van de Hulst (1949; curve No. 15) and for the JHKLMN passbands we follow the scaling laws provided by Rieke & Lebofsky (1985). We do allow for some random scatter around the average extinction. However, we do not consider (yet) the effects due to patchiness of the extinction along the line of sight. Ng & Bertelli (1996) demonstrated that this is in first approximation, visually almost indistinguishable from a random scatter around an average extinction.
In many studies the observations are crowding limited, due to the increasing number of stars towards fainter magnitudes. Crowding gives rise to star blends which affects the magnitude and the colour of the stars. The group of stars will be detected as a single star with a magnitude equal to the sum of the stellar flux of the stars involved in the stellar blend. The remaining stars are "hidden'' from detection.
The blends are well described as unresolved, apparent binaries. The simulation of apparent binaries is made with an iteratively improved blending probability, which is defined as the probability that a star within a given ensemble of stars might blend with another star from the same population. Each synthetic star within a stellar population is tested against the blending probability.
The percentage of artificial binaries is with this scheme about twice the blending factor. The blending factor in different passbands is not necessarily the same and the occurrence of star blends is furthermore not necessarily correlated, due to possible differences in the exposure time or seeing conditions.
The fainter companion stars of artificial binaries will give rise to incompleteness of the synthetic stellar sample. This allows us to map the synthetic stellar completeness function, which can be compared directly with the completeness function obtained from artificial star tests from the observed stellar sample.
In the CMD to be analysed we assume implicitly that,
between the observed and synthetic photometric system,
the uncertainty in the magnitude zeropoint is smaller than
the uncertainty in the zeropoint of the colour,
see Carraro et al. (1999), and references cited therein.
We allow for this reason the possibility that a small zeropoint
shift might be present between the colours of these systems.
![\begin{figure}
\par\includegraphics[width=8.2cm,clip]{aah3492f2a.eps}\hspace*{2mm}
\includegraphics[width=9.10cm,clip]{aah3492f2b.eps} %
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg30.gif) |
Figure 2: Schematic view of the main genetic operators acting on two sets of parameter strings in order to procreate two new sets (one chromosome set for each individual) of test strings: natural selection of two individuals G and g for the breeding of offspring, modification of the genetic content by means of a homologous crossover - at gene Gi/ gi, and finally the mutation of gene g2 to *g2. Each gene on a chromosome represent a parameter to be optimized. The schematics for a 2-point crossover scheme are shown for comparison |
| Open with DEXTER | |
Although straightforward, the generation of a large number of synthetic HRDs with each their own specific age-metallicity range, star formation history and initial mass function can be a time consuming task, because of the repetition of many calculations to generate one diagram. Figure 1 indicates that an alternative route with single stellar populations (SSPs) is available for the automated analysis. However, this method requires the computation of a large, regular grid of SSPs for different age-metallicity ranges and IMFs. The star formation history and age-metallicity range are the result of the linear combination of the SSPs. The time spent on computations of new CMDs thus can be greatly reduced through the use of probability density diagrams. However, this is not our prime objective. Our present goal is to develop an automated fitting method, which compares on a star by star basis, and to demonstrate its potential. Optimization for speed is not yet our primary concern. Moreover, the generation of one synthetic CMD with 5000 stars takes about 0.75 s on a PC equipped with a 200 MHz Pentium processor. This is a good indication that our present version of the software tool is performing at an acceptable speed. We refer to Dolphin (1997,2001,2002) and Olsen (1999) for a description of a method using SSPs.
| initial | PIKAIA | parameter | comment |
| value | default | identifier | |
| 100 | 100 | np | the number of individuals per population |
| 20 | 500 | ngen | number of generations |
| 2 | 5 | nd | number of digits encoding accuracy |
| 2 | 2 | imut | mutation mode, imut = 2 then pmut = [pmutmn,pmutmx] |
| 0.005 | 0.005 | pmut | initial mutation rate |
| 0.005 | 0.005 | pmutmn | minimum mutation rate |
| 0.35 | 0.25 | pmutmx | maximum mutation rate |
| 0.95 | 1.0 | fdif | fitness differential |
| 3 | 1 | irep | reproduction plan |
| 0 | 1 | ielite | elitism |
| 0 | 0 | ivrb | verbose mode |
The PIKAIA optimization package (V1.0; public domain) was developed by Charbonneau (1995) and a full description of this package is given by Charbonneauem&emKnapp (1996). PIKAIA has been used successfully in a wide range of astrophysical applications (e.g. Bobinger 2000; Charbonneau et al. 1998; Gibson & Charbonneau 1998; Kaastra et al. 1996; Kennelly et al. 1996; Lamontagne et al. 1996; McIntosh et al. 1998; Metcalfe 1999; Mewe et al. 1996; Noyes et al. 1997; Saha 1998; Wahde 1998).
PIKAIA is based on a genetic algorithm (Holland 1975; Goldberg 1989; Davis 1991; De Jong 1993), and is in principle not a function optimizer, but it does this extremely well. It searches for, locks on to, and pins down an optimal solution in a way, which is conceptually comparable to biological evolution through natural selection. Genetic algorithms are capable to explore and find in a robust way an optimum, but not necessarily the best, setting for a particular problem. In our case this comes down to minimizing the difference between a synthetic and an observed CMD by evolving the astrophysical parameters that define the shape of the CMD (see Table 2 and Sects. 2.10 and 3). We follow the generally accepted biological terminology for the description of a genetic algorithm.
A genetic algorithm makes use of a reduced version of the evolutionary process. The gene pool, i.e. the set of parameters to be optimized, and its associated phenotypic population evolves in response to
PIKAIA has 12 flow control parameters which are discussed
by Charbonneau (1995) to whom we refer
for a detailed description. We limit ourselves
to a short summary and the purposes of these control parameters:
np defines the number of individuals in a population;
ngen specifies the number of generations that the population
is evolving;
nd is number of digits encoding
accuracy![]() used for the parameters;
pcross is the probability that a crossover occurs between
the chromosomes of the parents;
imut,
pmut, pmutmn,
and pmutmx specify the mutation mode, the mutation rate
(the initial mutation rate if the rate spans the range from
pmutmn to pmutmx for
imutem=em2),
and the minimum and maximum mutation rate;
fdif the fitness differential controls the selection of
the individuals for breeding through their fitness;
irep defines the reproduction plan to be followed;
ielite defines if the fittest individual can or cannot
be selected for replacement; and
ivrb specifies verbose mode for extra on screen
information during the evolutionary run.
used for the parameters;
pcross is the probability that a crossover occurs between
the chromosomes of the parents;
imut,
pmut, pmutmn,
and pmutmx specify the mutation mode, the mutation rate
(the initial mutation rate if the rate spans the range from
pmutmn to pmutmx for
imutem=em2),
and the minimum and maximum mutation rate;
fdif the fitness differential controls the selection of
the individuals for breeding through their fitness;
irep defines the reproduction plan to be followed;
ielite defines if the fittest individual can or cannot
be selected for replacement; and
ivrb specifies verbose mode for extra on screen
information during the evolutionary run.
Table 1 holds a list of the PIKAIA flow parameters which were kept constant during all the simulations described in this paper. In the following subsections we describe the extensions added to the 12 flow parameters of PIKAIA.
The crossover operator is very effective in a global exploration of
the full parameter space and is in a way comparable to a variational
calculus method. A one-point crossover scheme, see
Fig. 2, is sometimes inadequate to combine and
pass on certain features encoded on the chromosomes (Michalewicz
1996) to its offspring. In some cases only a
correlated modification of a number of genes, say 2, will result in a
fitter offspring. This can, for example, be mimicked through the
application of a two- or multi-point crossover scheme. PIKAIA has
been extended with the control parameter rcross to handle a
multi-point crossover operation. For example: rcrossem=em1 represents the default one-point crossover,
while rcrossem=em2.3 represents a "2.3-point'' crossover:
i.e. a two-point and three-point crossover for respectively 70% and
30% of the cases (
![]() ).
).
An important drawback of genetic algorithms is that
the crossover operator is for about 75% of the time
lethal to its offspring, i.e. it produces children which
are not as fit as their parents (Banzhaf et al. 1998).
To avoid missing, potentially, fitter offspring and to
reduce the destructive effect of the crossover operator
we have incorporated brood recombination in PIKAIA
through a new control parameter rbrood.
The default PIKAIA reproduction scheme is obtained with
rbroodem=em1, i.e. two parents breed once and
produce two new individuals.
With
![]() the parents are allowed
to breed on average 3.5 times to produce a larger offspring (in this
case 7 on average).
For rbroodem>em1 one ends up with more
than two offspring. PIKAIA on the other hand expects from each pair
of parents only two children. This constraint was obeyed
in order not to significantly alter the global behaviour
of PIKAIA. Therefore, in order to avoid an exponential growth of
the population for rbroodem
the parents are allowed
to breed on average 3.5 times to produce a larger offspring (in this
case 7 on average).
For rbroodem>em1 one ends up with more
than two offspring. PIKAIA on the other hand expects from each pair
of parents only two children. This constraint was obeyed
in order not to significantly alter the global behaviour
of PIKAIA. Therefore, in order to avoid an exponential growth of
the population for rbroodem![]() em1,
only the fittest two individuals of the local offspring
survive
em1,
only the fittest two individuals of the local offspring
survive![]() and enter the global population for a fitness
evaluation from which a selection is made for further breeding.
The extra breeding increases the computational effort considerably,
due to a larger number of function evaluations. The advantage is a more
rapid increase of fitter individuals through the selection
of effective crossovers from good recombinations. On the other hand, a
rapid increase of fitter individuals might lead to a premature
convergence to a local minimum, due to a smaller variance in the genetic pool.
and enter the global population for a fitness
evaluation from which a selection is made for further breeding.
The extra breeding increases the computational effort considerably,
due to a larger number of function evaluations. The advantage is a more
rapid increase of fitter individuals through the selection
of effective crossovers from good recombinations. On the other hand, a
rapid increase of fitter individuals might lead to a premature
convergence to a local minimum, due to a smaller variance in the genetic pool.
We further introduced the "creep'' mutation (Charbonneau & Knapp 1996) in order to overcome the so-called "Hamming Wall'' problem, i.e. the inability to cross in a decimal encoding scheme certain boundaries with a one-point mutation operator. The creep parameter pcreep defines the probability that a gene in the pool undergoes a "standard'' mutation (change digit randomly in the range 0-9) or the "creep'' mutation (add or subtract one from the current value of the digit). We adopted as default equal weight for the occurrence of a "creep'' or "standard'' mutation.
We modified PIKAIA's uniform mutation mode. In the majority of the cases we require a (anti-)correlated change between two or more parameters (see Sect. 6.2). In a standard mutation scheme convergence might be slow if one has to wait for the simultaneous occurrence of a favourable (anti-)correlated mutation of two specific parameters in order to improve the fitness. We introduced an extra parameter pcorr which defines the probability that a correlated mutation occurs. If this is not the case the standard mutation scheme is chosen. Otherwise we allowed that in 50% of the cases the mutations of two genes (igen1 and igen2 are extra input parameters added to the modified version of PIKAIA) are more relevant than the mutations occurring in other parameters. For the remaining 50% of the cases the two genes are determined stochastically. Additional details about the adopted values of the control parameters are given in Sect. 2.10 and Table A.1.
![]() and
and ![]() are respectively defined as:
are respectively defined as:
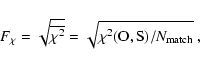 |
(3) |
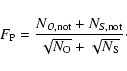 |
(4) |
| # | parameter | value | lower limit | upper limit | description |
| 1 | log d | 3.7 | 3.6 | 4.0 | log of distance |
| 2 | 0.30 | - 0.001 | 0.5 | extinction | |
| 3 |
|
9.5 | 9.0 | 10.3 | log lower age limit |
| 4 |
|
- 0.90 | - 1.69897 | 0.69897 | log lower metallicity limit |
| 5 | 1.35 | 1.001 | 3.5 | IMF slope | |
| 6 |
|
9.6 | 9.0 | 10.3 | log upper age limit |
| 7 | [Z]
|
0.30 | - 1.69897 | 0.69897 | log upper metallicity limit |
| 8 | 1.0 | - 2.00 | 5.00 | SFR slope |
The formal 1![]() uncertainty of each parameter k,
see Table 4,
is obtained through variation of this parameter and
by minimizing the function
uncertainty of each parameter k,
see Table 4,
is obtained through variation of this parameter and
by minimizing the function
![]() .
Conceptually, this is similar to moving the merit function
in the
.
Conceptually, this is similar to moving the merit function
in the
![]() - plane away from its optimum setting,
to the nearest position on a contour +1
- plane away from its optimum setting,
to the nearest position on a contour +1![]() higher.
The associated fitness
function
higher.
The associated fitness
function
![]()
![]() is:
is:
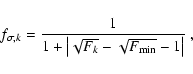 |
(5) |
We implemented a hybrid optimizer in which we use PIKAIA to explore the parameter space and then use POWELL's minimization algorithm (Powell 1964; Press et al. 1986) to pin down the nearest local or global minimum through a direction set method which produces N mutually conjugate (non-interfering) directions. For details and an excellent description of this algorithm we refer to Press et al. and references cited therein.
A hybrid minimization strategy is used, because PIKAIA is by definition not a function optimizer, but it tends to get close near a (local) optimum. POWELL is used to get even closer to the (local) optimum. If we had landed in a local optimum then we needed PIKAIA to jump out of it. The origin of our need for a hybrid search strategy is comparable to the minimization problems encountered by Harris & Zaritsky (2001).
We implemented a dynamic, scalable parameter range in our search for an optimum set of parameters. The parameter range shrinks after each optimization cycle with POWELL. This leads to an improved accuracy in the results with a fixed number of digits encoding accuracy.
The automated re-scaling of the parameter range improves the resolution of the exploration of the search grid. In addition, due to the re-scaling one may circumvent partial degeneracy of the parameters.
The global function
![]() gives the global distance to the minimum
in terms of
gives the global distance to the minimum
in terms of
![]() .
If we have n parameters then each parameter k is in a simple approach
on average about
.
If we have n parameters then each parameter k is in a simple approach
on average about
![]() away from its optimum
value,
because we assume that
away from its optimum
value,
because we assume that
AMORE uses in essence two different sets of parameters as input. One set contains program flow control parameters and information about the observational data to be simulated: the photometric errors and crowding factors per passband, a shift of the zeropoint of the colours due to a difference between the observed and synthetic photometric system, and the spread around the average extinction value to account partly for the differential reddening in a field.
![\begin{figure}
\par\includegraphics[width=10.5cm,clip]{aah3492f3.eps}
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg70.gif) |
Figure 3:
Evolutionary status for all the trial models
(filled dots; see Table A.1
for details) for each population after 400 generations, see also
Fig. 4.
The filled star
|
| Open with DEXTER | |
The other set of input parameters is used by PIKAIA and can be divided into two parts (see Tables 1 and 2). One part contains the PIKAIA control parameters. The other part contains the lower and upper limits of the astrophysical parameters to be optimized as well as an initial guess for the value of those parameters (first column of Table 2, resulting HRD in Fig. 6b). These parameters are a combination of the synthetic population's intrinsic properties, i.e. age, metallicity, slope of the power-law IMF and the index of the exponential SFR. In our case we allow for age and metallicity not to be restricted to one fixed value, but to cover a specific range (see Table 2).
In addition, there are two parameters which mimic the synthetic population's behaviour as placed in a mockup version of our Galaxy. These parameters are the distance from the Sun and the average extinction.
In total we thus have 8 free parameters which AMORE has to optimize simultaneously.
In terms of genetic programming the objective of AMORE is to determine
the genome (i.e. the set of astrophysical parameters
described in Sect. 2.10, see also Table 2)
of a specified individual (i.e. the observed stellar population).
Note that it is not possible to directly observe the genome.
The genome is determined from the phenotype of each individual
(i.e. the synthetic CMDs, see for example
Fig. 6). The genetic information is located
in the genes of one chromosome![]() .
.
![\begin{figure}
\par\includegraphics[width=18cm,clip]{aah3492f4.eps}
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg71.gif) |
Figure 4: The filled circles in panels a)- f) display the values obtained for the parameters using the models in Table A.1. The solid line refers to the value set for the original population. |
| Open with DEXTER | |
A guess of the genotype of the observed CMD is obtained
through comparison with a synthetic CMD, which is generated
via supervised evolution and breeding
(PIKAIA together with POWELL).
The stars in the synthetic CMD population with a particular genotype
are raised to maturity (HRD-ZVAR and HRD-GST).
A group of individuals![]() is allowed to procreate (the chance of an
individual procreating depends on its fitness and the selection
pressure, see Charbonneau 1995 and Charbonneau & Knapp
1996) and the genetic information of the parents is passed on
to their offspring (see Fig. 2).
A fitness evaluation
(a comparison between the observed and synthetic CMD)
provides a ranking of the resulting group of individuals.
If the individual has "good genes''
it survives, remains in the group
and gets a chance of procreation.
is allowed to procreate (the chance of an
individual procreating depends on its fitness and the selection
pressure, see Charbonneau 1995 and Charbonneau & Knapp
1996) and the genetic information of the parents is passed on
to their offspring (see Fig. 2).
A fitness evaluation
(a comparison between the observed and synthetic CMD)
provides a ranking of the resulting group of individuals.
If the individual has "good genes''
it survives, remains in the group
and gets a chance of procreation.
The evolutionary process of breeding and fitness evaluation is repeated for a fixed number of generations. The gene pool of the resulting best individual at the end of the evolutionary run with AMORE hopefully represents a near-optimum representation of the unknown genome.
Initially PIKAIA is in control (see Fig. 1) of the evolution for a fixed number of generations. Afterwards POWELL tries to improve the genome of the fittest individual communicated through PIKAIA. We then determine the uncertainty for each gene on the chromosome. Subsequently, we tighten the limits on the range of variation allowed for each gene and re-scale the parameters on the genetic print of the fittest individual accordingly. During the shrinkage of the parameter range we do not re-scale the genetic information of the remaining individuals, but preserve their former values as semi-random input for the continued optimization process. The latter addition to the hybrid scheme is most likely a significant driver in speeding up the search for a fitter individual.
After each optimization with POWELL a new cycle with PIKAIA is started with the current best parameter set as "educated next guess'' for AMORE's progressive evolution. The total number of PIKAIA cycles is user defined.
In Sect. 2.9 we argued that
the parameters are on average about
![]() away
from its optimum value.
The convergence however is not governed by the average "distance''
that each parameters is away from its optimum setting. It is mainly
determined from the ability to tune the parameter which has
the largest offset from its optimum value.
away
from its optimum value.
The convergence however is not governed by the average "distance''
that each parameters is away from its optimum setting. It is mainly
determined from the ability to tune the parameter which has
the largest offset from its optimum value.
In the AMORE training sessions it was noted that with
![]() about three of the eight
parameters are about 1em
about three of the eight
parameters are about 1em![]() (
(
![]() )
away from their optimum value.
AMORE
has a built-in option to do a random variation from 0-3
)
away from their optimum value.
AMORE
has a built-in option to do a random variation from 0-3 ![]() of
two parameters
(randomly selected)
from the
running best solution when the fitness is less than 0.30.
Above this threshold we choose a new value
for two parameters according
of
two parameters
(randomly selected)
from the
running best solution when the fitness is less than 0.30.
Above this threshold we choose a new value
for two parameters according
![]() ,
where
,
where
![]() can be any number between 1.0 and 4.0.
can be any number between 1.0 and 4.0.
We explored several different settings for the PIKAIA control parameters, because the tuning of those parameters is very problem dependent (Charbonneau & Knapp 1996). The values we decided to use are listed in Table 1. Four notes can be made here.
Firstly the steady-state-delete-worst reproduction plan (irep=3) we adopted, in which we replace the least-fit individual from the population when the fitness of the new individual is superior to that of the least-fit population member. Choosing this reproduction plan implies that the elitism control parameter (ielite) is non-operative, because elitism is active by default. We evaluated two other reproduction plans (Charbonneau & Knapp 1996); full generational replacement and steady-state-delete-random. The steady-state-delete-worst reproduction plan produced on average the best results.
Secondly the mutation rate of 0.35 corresponds, in case of a default 2 digit accuracy, with the on average occurrence of 2.8 mutations per astrophysical parameter.
Thirdly, the fitness differential parameter fdif, a measure for the selection pressure, would normally be chosen as high as possible (fdif=1 in this case). However, it may possible to circumvent local minima by lowering that value a bit (Charbonneau & Knapp 1996). Setting fdif=0.95 turned out to be a good trade-off choice.
Fourthly, we want to explore as large a fraction of the parameter space as possible at the first entry in AMORE. This is done by using only an one digit accuracy (nd=1). Due to the active re-scaling of the parameter space boundaries we do not require a very high precision in our exploration. A one percent accuracy (nd=2) of the parameter space is sufficient in the subsequent PIKAIA cycles.
In biological terms, the PIKAIA control parameters define the ecosystem in which our population evolves.
All computations presented in this paper were performed
with an executable generated with the g77 compiler.
This executable was then installed on
various PCs running Red Hat
Linux![]() 6.X and 7.X.
The PCs were equipped with Intel Pentium III or Athlon processors
with clock speeds ranging from 600 - 1200 MHz.
6.X and 7.X.
The PCs were equipped with Intel Pentium III or Athlon processors
with clock speeds ranging from 600 - 1200 MHz.
All tests, unless stated otherwise, use the synthetic population as described by Ng (1998):
| parameter | value |
|
|
|
|
| pcross | 0.50 | 0.276 | 0.070 | 0.287 | 0.066 |
| 0.85 | 0.299 | 0.063 | 0.297 | 0.056 | |
| rcross | 1.00 | 0.293 | 0.065 | 0.290 | 0.063 |
| 2.00 | 0.285 | 0.068 | 0.297 | 0.065 | |
| 3.00 | 0.285 | 0.070 | 0.290 | 0.057 | |
| rbrood | 1.00 | 0.286 | 0.077 | 0.230 | 0.059 |
| 2.00 | 0.292 | 0.058 | 0.301 | 0.055 | |
| 4.00 | 0.285 | 0.066 | 0.276 | 0.067 | |
| pcreep | 0.0 | 0.287 | 0.071 | 0.292 | 0.068 |
| 0.3 | 0.290 | 0.065 | 0.297 | 0.058 | |
| 0.7 | 0.285 | 0.066 | 0.288 | 0.060 | |
| pcorr | 0.0 | 0.278 | 0.077 | NA | NA |
| 0.3 | 0.285 | 0.065 | NA | NA | |
| 0.7 | 0.300 | 0.057 | NA | NA |
In the first test, we evaluate the 162 models listed in Table A.1 in order to study the effect of the PIKAIA parameters pcross, rcross, rbrood, pcreep, and pcorr on the convergence and computational effort. The test has as a secondary objective to provide an understanding of the degeneracy of the parameter space.
All astrophysical parameters to be retrieved are set free, floating
between reasonable minimum and maximum values (see
Table 2 for details). AMORE runs for 20 iterations of 20 generations (ngen=20) to recover the a priori known parameters of the synthetic population.
The number of iterations and generations
determine the total length of an evolutionary run:
![]() generations.
Note that the
range of each parameter is set within reasonable limits and not taken
excessively large, because it might lead to the case that no
acceptable parameter setting is found with the standard iteration loop.
generations.
Note that the
range of each parameter is set within reasonable limits and not taken
excessively large, because it might lead to the case that no
acceptable parameter setting is found with the standard iteration loop.
| parameter | log d(pc) |
|
|
|
|
|
|||
| original | 3.906335 | 0
|
9.90309 | 9.95424 | - 0.60206 | 0.17609 | 2.35 | 1.0 | 0.44597 |
| round-v1 | 3.906 | 0
|
9.903 | 9.954 | - 0.60 | 0.18 | 2.35 | 1.0 | 0.28595 |
| round-v2 | 3.906 | 0
|
9.903 | 9.954 | - 0.602 | 0.176 | 2.35 | 1.0 | 0.30812 |
| round-v3 | 3.9063 | 0
|
9.9031 | 9.9542 | - 0.602 | 0.176 | 2.35 | 1.0 | 0.42439 |
| average value | 3.8958 | 0
|
9.866 | 9.984 | - 0.554 | 0.244 | 2.358 | 1.574 | |
| 0.0033 | 0
|
0.049 | 0.047 | 0.050 | 0.13 | 0.034 | 1.40 | ||
|
|
0.012 | 0
|
0.043 | 0.023 | 0.18 | 0.08 | 0.03 | 1.4 |
In the third test we take six models in which one of the parameters is set fixed at its correct value in order to study the effects on the convergence. The models chosen were two of high, two of intermediate and two of low fitness as determined from the first test. The convergence in this test basically can go two ways: either the convergence is faster, because less parameters have to be optimized. Or, due to the fact that AMORE has less maneuverability in this situation, the convergence is slower. We adjusted the limits for age and metallicity as given in Table 2 such that AMORE would not try to find solutions in forbidden regions of parameter space which might severely slow down convergence due to constant rejection by AMORE of the chosen parameter values.
For example, fixing the
![]() parameter at its correct value of - 0.60206 means that we have to adjust the lower limit for
parameter at its correct value of - 0.60206 means that we have to adjust the lower limit for
![]() to - 0.60206 as well.
to - 0.60206 as well.
In the case of fixing the
![]() parameter this also
implies that the initial guess has to be adjusted. We set this initial
guess to 10.1.
parameter this also
implies that the initial guess has to be adjusted. We set this initial
guess to 10.1.
In the fourth test we take six models in which one of
the parameters is set fixed at 1![]() offset
(determined
from the first test) from its original value, in order to
study its effect on the "second best'' setting of the remaining parameters.
Normally one would expect a fitness
offset
(determined
from the first test) from its original value, in order to
study its effect on the "second best'' setting of the remaining parameters.
Normally one would expect a fitness
![]() .
In this case, however,
.
In this case, however,
![]() and the
associated fitness constraint drops to
and the
associated fitness constraint drops to
![]() .
However, this assessment ignores the fact that, when a parameter is
offset from its optimum value, the number of matched points will
decrease and
.
However, this assessment ignores the fact that, when a parameter is
offset from its optimum value, the number of matched points will
decrease and ![]() increases. Using Eq. (6)
one has for a good fit
increases. Using Eq. (6)
one has for a good fit ![]() .
On average the offset
per parameter k from the optimum value is
.
On average the offset
per parameter k from the optimum value is
![]() ,
at best
the offset is
,
at best
the offset is
![]() ,
and in the worst case this is
,
and in the worst case this is
![]() .
So with one parameter k put at
.
So with one parameter k put at
![]() offset we
distinguish the three possibilities
offset we
distinguish the three possibilities
| 1 | at best |
|
=3 |
|
|
| 2 | on average |
|
=4 |
|
|
| 3 | at worst |
|
=5.8 |
|
|
The effect of the 1em![]() offset of one of the parameters
will partly be canceled by forcing other parameters away
from the optimum value. For example, the effect of an increased
extinction can be masked partially by generating a bluer
stellar population with a lower metallicity and a younger age.
The effect will be such that the fitness will not
be around
offset of one of the parameters
will partly be canceled by forcing other parameters away
from the optimum value. For example, the effect of an increased
extinction can be masked partially by generating a bluer
stellar population with a lower metallicity and a younger age.
The effect will be such that the fitness will not
be around
![]() ,
but
somewhere in the range
,
but
somewhere in the range
![]()
We fixed the parameters both at one sigma above and one sigma below the original value, because the evolutionary effects do not have to be symmetric. The only exception is the extinction, which we only fix at one sigma above the original value of AV = 0.0.
Again we adjusted the limits for the upper and lower limit for age and metallicity.
To better understand what goes on during the genetic evolution we display the results from model A.1-40. Figure 5 displays an example of the evolution of the merit function F for a number of generations. It shows how the initially dispersed individuals gradually find their way, start to cluster together around generation 10, and penetrate the region with acceptable solutions after about 50 generations. After 100 generations the improvements become marginal for this model.
Figure 6 displays for the same model A.1-40 the phenotypical changes of the CMD for several fitnesses during the genetic evolution. The various panels show that the synthetic CMD resembles better and better the "observed'' CMD when the fitness improves. Note that at fitness fem=em0.05 one already gets for the eye appealing solutions.
Figure 7 shows the improvements of the
astrophysical parameters as a function of increasing fitness
for the models A.1-40 and A.1-51.
The panels for distance and extinction show that the
distance is systematically underestimated, while the
extinction is overestimated. But in general one notices that
the astrophysical parameters obtained from model
A.1-40 get quite close to the parameters
of the CMD to be matched.
![\begin{figure}
\par\includegraphics[width=18cm,clip]{aah3492f5.eps}
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg100.gif) |
Figure 5:
Conception diagram of the
evolution of the genetic population of model A.1-40 during
the optimization process displayed in Fig. 7.
Frame a) shows the initial population and the frames
b)- l) show the population after several generations
up to generation = 400. The
outer
shaded region indicates solutions for which
the difference between the CMDs from the "observed'' and synthetic
population is on average
between 1-3em |
| Open with DEXTER | |
Table A.1 is displayed in
Fig. 3. The clustering in the figure provides
an indication that a degeneracy of the parameter space is present near
![]() (i.e.
(i.e. ![]() ,
see Eq. (1)).
Without a major computational effort
it will be difficult to obtain a significant improvement of the
parameters once
,
see Eq. (1)).
Without a major computational effort
it will be difficult to obtain a significant improvement of the
parameters once
![]() .
However,
.
However, ![]() indicates a region in the (
indicates a region in the (
![]() )-plane
for which the
systematic offset of the individual parameters
from its true value
are on average less than
)-plane
for which the
systematic offset of the individual parameters
from its true value
are on average less than
![]() ,
see Sect. 2.9 for details.
In practice it turns out that
a strong correlation between three of the eight parameters
has the culprit; at least two of them have to change simultaneously
in the proper direction in order to improve the fitness
(see also Sect. 6.3).
They have an
average offset of
,
see Sect. 2.9 for details.
In practice it turns out that
a strong correlation between three of the eight parameters
has the culprit; at least two of them have to change simultaneously
in the proper direction in order to improve the fitness
(see also Sect. 6.3).
They have an
average offset of
![]() ,
while for the remaining parameters
this is
,
while for the remaining parameters
this is ![]()
![]() .
.
In addition, Fig. 4 displays the retrieved
parameters for all models
as a function of fitness. Note, that AMORE systematically
underestimates the distance of the test population. On the other hand,
the effect of this underestimation is in its turn partially canceled by
overestimating the extinction, the upper
age limit and the slope of the power-law IMF slightly
(see also Fig. 7).
Another clue we get from Fig. 4 is that the slope
of the SFR ![]() is very poorly constrained.
is very poorly constrained.
![\begin{figure}
\par\mbox{\hspace{5cm}\includegraphics[width=5cm,clip]{aa3492f6a....
...ps}\hspace{2mm}
\includegraphics[width=5cm,clip]{aa3492f6j.eps} }\end{figure}](/articles/aa/full/2002/36/aah3492/Timg107.gif) |
Figure 6:
Genetic evolution of the colour-magnitude diagram (CMD)
from the first test population.
Panel a) displays the original population to be matched.
The physical parameters for this population are described
Table 2 and Sect. 4.2. The CMD of the initial trial
population is shown in panel b). Panels c)- e) display the
resulting CMDs obtained with setup A.1-40 for different
fitnesses (see Sect 2.7). The fitnesses f = 0.05, f = 0.19and f = 0.27 are respectively reached after 20, 60 and 80 generations.
The dots in the panels b)- e) are used for each matching point,
while the red open stars
|
| Open with DEXTER | |
![\begin{figure}
\par\includegraphics[width=18cm,clip]{ONLINE_FILES/A_SCANNER/evolve.ps}
\end{figure}](/articles/aa/full/2002/36/aah3492/Timg109.gif) |
Figure 7:
Panels a)- h) display the
convergence curves for the parameters of models A.1-40 (dotted line; fitness f=0.41)
and A.1-51 (long dashed line; fitness f=0.25).
The solid line in the frames a)- f) refers
to the value adopted for the original population.
The long, dot dashed line in frames i) and j)
shows the threshold values to be crossed for acceptable solutions,
i.e. F < 2 and
|
| Open with DEXTER | |
In order to compensate for this strong stabilizing effect, we also evaluate in Table 3 the average fitness of the models when we exclude all models which have pcorr = 0.0.
As expected, the rbrood parameter has a strong influence on the
amount of computational time needed. Although the models
with high values of rbrood are somewhat better than
models with low values, this effect is only marginal. Considering that a
high value of rbrood lessens the genetic variation in the gene
pool while increasing the computational time needed for a run with
several factors, it is desirable to have a low value of rbrood.
The different parameters are not independent, as can be seen from
Table A.1 and Table 3.
Simply taking the best options in Table 3 yields
model 134 for the case in which pcorr = 0.0 has not been corrected
for, a reasonable, but not an exceptionally good model.
The true ![]() line in the table shows that both the
line in the table shows that both the
![]() and the
and the ![]() parameter are the weak links in the overall
parameter estimation (see also Fig 4).
parameter are the weak links in the overall
parameter estimation (see also Fig 4).
| parameter |
|
|
| log d | 0.299 | 0.081 |
| 0.274 | 0.081 | |
|
|
0.332 | 0.021 |
|
|
0.304 | 0.056 |
|
|
0.269 | 0.036 |
|
|
0.361 | 0.036 |
| 0.305 | 0.066 | |
| 0.312 | 0.076 |
The results of fixing parameters at the correct value are listed in Table A.2 and an example of the diagnostics is listed in Table 6. Details of the individual setups for these tests are given below. In general, the results of the tests for which one of the parameters was set to the correct value were slightly better than the results for the models for which all parameters are set free, see Tables 4 and 5 for additional details. This behaviour is due to the fact that by forcing one parameter to a fixed value the evolutionary path changes. The models were selected from the results with low and intermediate fitness given in Table A.1.
The lower value for the
average fitness
![]() ,
when fixing the distance at the correct value,
is caused by the presence of
one outlier (see Table A.2),
which is caused by the age-metallicity degeneracy.
Excluding this value results in
an average fitness of
,
when fixing the distance at the correct value,
is caused by the presence of
one outlier (see Table A.2),
which is caused by the age-metallicity degeneracy.
Excluding this value results in
an average fitness of
![]() .
In general: the extinction
can be reliably retrieved when fixing the distance.
.
In general: the extinction
can be reliably retrieved when fixing the distance.
When considering a fixed extinction, the results show a strong variation in
both age and metallicity. It should also be noted that the average
![]() em(distance (pc))
retrieved is only 3.8953em
em(distance (pc))
retrieved is only 3.8953em![]() em0.0066.
This is more than one sigma away from the optimum value for the distance (see
Table 4). This is an indication that retrieval of the
distance by fixing the extinction is hampered by
the age-metallicity degeneracy. Therefore, the distance cannot be
reliably retrieved when fixing the extinction to its correct value.
em0.0066.
This is more than one sigma away from the optimum value for the distance (see
Table 4). This is an indication that retrieval of the
distance by fixing the extinction is hampered by
the age-metallicity degeneracy. Therefore, the distance cannot be
reliably retrieved when fixing the extinction to its correct value.
Fixing one of the age limits results in values for both the age and metallicity which are close to the input values. This is due to the (partial) breaking of the age-metallicity degeneracy. The distance-extinction degeneracy remains. The results also suggest that the age-metallicity degeneracy has a stronger impact on the fitness than the distance-extinction degeneracy.
Fixing the upper metallicity limit to its
correct value shows that the values for age and metallicity
come closer to their original, input values.
This is quite in
contrast with the results obtained from fixing the
lower metallicity to its correct value.
Table A.2 shows that both the high metallicity
limit and the slope of the exponential SFR are not well
constrained. This behaviour can be accounted to the
implicit shape of the linear age-metallicity relation
adopted in the HRD-GST. The number of high metallicity
stars is smaller than the number of low metallicity
stars due to the adopted, exponentially decreasing (![]() ),
star formation rate. The consequence is that the high metallicity
limit can be better determined when both the low and high
limits are determined in union.
),
star formation rate. The consequence is that the high metallicity
limit can be better determined when both the low and high
limits are determined in union.
| model |
|
|
|
F | |||
| ideal | 0 | 0 | 5000 | 0.000 | 0.000 | 0.000 | 1.000 |
| free | 60 | 60 | 4940 | 0.847 | 0.849 | 1.438 | 0.410 |
| fixed: log d (pc) | 155 | 155 | 4845 | 0.903 | 2.192 | 5.621 | 0.151 |
| fixed: |
103 | 103 | 4897 | 0.840 | 1.457 | 2.827 | 0.261 |
| fixed:
|
76 | 76 | 4924 | 0.817 | 1.075 | 1.823 | 0.354 |
| fixed:
|
104 | 104 | 4896 | 0.902 | 1.471 | 2.978 | 0.251 |
| fixed:
|
94 | 94 | 4906 | 0.836 | 1.329 | 2.466 | 0.289 |
| fixed:
|
76 | 76 | 4924 | 0.815 | 1.075 | 1.820 | 0.355 |
| fixed: |
74 | 74 | 4926 | 0.834 | 1.047 | 1.799 | 0.357 |
| fixed: |
80 | 80 | 4920 | 0.727 | 1.131 | 1.808 | 0.356 |
| parameter | offset |
|
|
| log d | 0.291 | 0.048 | |
| 0.280 | 0.066 | ||
| 0.284 | 0.020 | ||
|
|
0.258 | 0.044 | |
| 0.279 | 0.012 | ||
|
|
0.229 | 0.022 | |
| 0.207 | 0.014 | ||
|
|
0.254 | 0.066 | |
| 0.315 | 0.027 | ||
|
|
0.270 | 0.054 | |
| 0.283 | 0.018 | ||
| 0.266 | 0.025 | ||
| 0.273 | 0.027 | ||
| 0.232 | 0.071 | ||
| 0.276 | 0.011 |
The results of fixing the parameters at a
![]() offset from its original value are listed in Table A.3.
An example of the diagnostics of these tests are given in
Table 8. Details of individual setups are given below.
offset from its original value are listed in Table A.3.
An example of the diagnostics of these tests are given in
Table 8. Details of individual setups are given below.
If the distance is overestimated there are more synthetic stars present at fainter magnitudes. To get relatively more synthetic stars at brighter magnitudes one needs to flatten the power-law IMF slope.
If the distance is underestimated then more synthetic stars are present at brighter magnitudes. One expects that a steeper power-law IMF slope is required as compensation. This is not always true, see Table A.3. Stars pop up at the lower end of the main sequence. They are taken away from the stars located at brighter and brighter magnitudes. One therefore requires also in this case a flatter IMF slope.
A flatter slope of the power-law IMF can be a hint that the distance of the stellar aggregate is wrong. Or it might be a hint that the zero point of the adopted synthetic photometric system is different from the actual photometric system used.
Recognizing that the slope is indeed flatter than the majority of the other cases outlined in Table 4 one may start to explore the assumption that the distance is wrong: release the constraint during the next exploration.
The sensitivity to the distance implies that AMORE can be used to determine the distance to a stellar aggregate quite reliably. A bonus is that due to an initially wrongly assumed distance the extinction is in most cases better constrained.
A higher value of the star formation index results in a lower number of stars at the upper age metallicity limit. To get a sufficient number of high metallicity stars one has to stretch the upper metallicity limit to a slightly higher value.
We further notice that a wrong value for the age and metallicity does not
affect the extinction significantly.
Our findings indirectly supports the method to determine
high resolution (
![]() )
extinction maps towards the Galactic bulge by Schultheis
et al. (1999) with the data obtained for the DeNIS project
(Epchtein et al. 1997).
)
extinction maps towards the Galactic bulge by Schultheis
et al. (1999) with the data obtained for the DeNIS project
(Epchtein et al. 1997).
The slope of the power-law IMF is
very strongly constrained (Ng 1998) for the test population.
As a consequence the changes in the values of the
remaining parameters are not extremely large.
The slightly larger value of the slope ![]() pushes slightly more synthetic stars to fainter magnitudes,
introducing a relative deficiency of stars at brighter magnitudes.
This is compensated through a younger age, a decrease
of the lower metallicity limit and an increase of the upper
metallicity limit. The higher value for the upper age
and metallicity limit compensates
in its turn for the overestimation of the exponential
star formation index.
pushes slightly more synthetic stars to fainter magnitudes,
introducing a relative deficiency of stars at brighter magnitudes.
This is compensated through a younger age, a decrease
of the lower metallicity limit and an increase of the upper
metallicity limit. The higher value for the upper age
and metallicity limit compensates
in its turn for the overestimation of the exponential
star formation index.
A lower value of ![]() is partly compensated for by lowering the
upper age limit, lowering the lower metallicity limit and
overestimating the upper metallicity limit.
is partly compensated for by lowering the
upper age limit, lowering the lower metallicity limit and
overestimating the upper metallicity limit.
A larger index for an exponentially decreasing SFR pushes more stars of the population to the blue edge of the CMD, resulting in a slightly bluer stellar population. AMORE compensates this by mainly increasing the upper metallicity limit, i.e. reddening the synthetic stellar population.
A lower value for the SFR index has the opposite effect. AMORE compensates for the now slightly redder population by lowering the upper metallicity limit, making the population bluer.
| model | offset |
|
|
|
F | |||
| ideal | 0 | 0 | 5000 | 0.000 | 0.000 | 0.000 | 1.000 | |
| free | 60 | 60 | 4940 | 0.847 | 0.849 | 1.438 | 0.410 | |
| fixed: log d (pc) | 91 | 91 | 4909 | 0.788 | 1.287 | 2.278 | 0.305 | |
| 85 | 85 | 4915 | 0.722 | 1.202 | 1.967 | 0.337 | ||
| fixed: |
98 | 98 | 4902 | 0.932 | 1.386 | 3.790 | 0.264 | |
| fixed:
|
91 | 91 | 4901 | 0.897 | 1.287 | 2.460 | 0.289 | |
| 94 | 94 | 4906 | 0.905 | 1.329 | 2.587 | 0.278 | ||
| fixed:
|
106 | 106 | 4894 | 0.977 | 1.499 | 3.202 | 0.238 | |
| 126 | 126 | 4874 | 1.017 | 1.782 | 4.211 | 0.192 | ||
| fixed:
|
78 | 78 | 4922 | 0.907 | 1.103 | 2.043 | 0.329 | |
| 80 | 80 | 4920 | 0.835 | 1.131 | 1.977 | 0.336 | ||
| fixed:
|
148 | 148 | 4852 | 0.896 | 2.094 | 5.188 | 0.162 | |
| 94 | 94 | 4906 | 0.867 | 1.329 | 2.519 | 0.284 | ||
| fixed: |
95 | 95 | 4905 | 0.886 | 1.344 | 2.590 | 0.279 | |
| 103 | 103 | 4897 | 0.894 | 1.457 | 2.921 | 0.255 | ||
| fixed: |
145 | 145 | 4855 | 0.930 | 2.051 | 5.071 | 0.165 | |
| 103 | 103 | 4897 | 0.834 | 1.457 | 2.818 | 0.262 |
The origin of this behaviour lies in the implicit definition of the
exponential star formation rate (for
![]() one has a decreasing star
formation towards a younger age) attached to a linear age-metallicity
relation. The latter relation will give less metal-richer stars.
The small number of stars with higher metallicity induces a larger
variation of the
one has a decreasing star
formation towards a younger age) attached to a linear age-metallicity
relation. The latter relation will give less metal-richer stars.
The small number of stars with higher metallicity induces a larger
variation of the
![]() parameter without affecting significantly
the overall fitness.
parameter without affecting significantly
the overall fitness.
We did not want to deal with a mutation dominated search, because it tends to move farther away from an optimum parameter setting in the majority of the cases. We used therefore a relatively high crossover probability (pcross) and we set the mutations at a fixed rate, such that on average only 2.8 mutations occur in the gene pool of each individual.
At a certain stage however one requires the variation of other correlated parameters to obtain an improvement. This becomes particularly necessary when approaching the optimum setting of the parameters. A favourable crossover and mutation might do the trick, but it might take a while before this occurs. We introduced in Sect. 2.6.5 the possibility that two parameters might be more sensitive to mutations than others. This approach gave better results for the majority of the trial cases (see Table 3), but it failed to obtain improvements when changes of one parameter were neutralized through the variation of one or more parameters. The distance-extinction and the age-metallicity degeneracies slow down the convergence of AMORE for f>0.3, see Fig. 4.
One of the modifications to consider for future implementation
is a two-chromosome approach. In that case
acceptable values for the parameters do not shift out of the
population if the overall fitness is less, but still
reside in the gene pool as a recessive quality. This however, will
require a major extension to PIKAIA and a significant amount
of genetic research to be done about dominant and recessive qualities
in the AMORE gene pool.
Another modification to consider in order to improve the accuracy and
to speed up convergence, is to replace the finite resolution of the
digital encoding scheme with a genetic coding based on floating point,
i.e. each gene on the chromosome is represented by one floating point
number. According to Michalewicz (1996) a real encoding
scheme can be superior and improve convergence.
Such an encoding scheme is indeed to be included in the
next release of PIKAIA 2.0 (Charbonneau; in preparation).
As demonstrated in Sect. 5.1.1 the degeneracy among parameters
becomes noticeable for f > 0.25 or F < 3.
This corresponds to
systematic offset
for each parameter
of on average ![]()
![]() and at maximum
and at maximum ![]()
![]() .
The Poisson uncertainty of the original population results in
a fitness of
.
The Poisson uncertainty of the original population results in
a fitness of
![]() (
(
![]() ).
However, solutions with a comparable fitness do exist due
to the degeneracy of the parameter space.
A direct consequence is that there is an intrinsic
offset
present among the parameters amounting to
on average
).
However, solutions with a comparable fitness do exist due
to the degeneracy of the parameter space.
A direct consequence is that there is an intrinsic
offset
present among the parameters amounting to
on average ![]()
![]() and at maximum
and at maximum ![]()
![]() .
This intrinsic
offset
is present in the solutions obtained
with AMORE and actually is responsible for slowing down the convergence
in the fitness range
0.30<f<0.43.
It will therefore be nearly impossible to
recover in one pass the original input values.
However, some improvements might be obtained by averaging the parameter
values obtained from AMORE runs with different initial conditions.
.
This intrinsic
offset
is present in the solutions obtained
with AMORE and actually is responsible for slowing down the convergence
in the fitness range
0.30<f<0.43.
It will therefore be nearly impossible to
recover in one pass the original input values.
However, some improvements might be obtained by averaging the parameter
values obtained from AMORE runs with different initial conditions.
Despite limitations in the input physics of the underlying stellar evolutionary tracks and by the transformation from the theoretical to the observational plane, the results with real data from Gallart et al. (1999), who uses the same set of evolutionary tracks, are encouraging. It will therefore be important to verify first with, for example, well studied open clusters (see Carraro et al. 1998,1999 and references cited therein) for which age and metallicity range we may apply AMORE safely. Extinction is also of some concern, because a high extinction may result in the MS turnoff point to fall below the detection limit. This would deprive AMORE of a clear reference point.
Another case of interest is of course the question how many different stellar populations can be distinguished with AMORE. Separating multiple, mixed populations from each other through the automated and the objective analysis of colour-magnitude diagrams could be a valuable tool for the analysis of galaxy formation and evolution.
This requires a rigorous follow up study on the separation of multiple (synthetic) populations. One further ought to verify if the automated analysis of colour-colour diagram can reduce the effects of error cancellation between distance and extinction.
Finally, after a succesful implementation, testing and validation phase, we plan to combine AMORE with the Padova spectrophotometric code (see Bressan et al. 1994, 1996; Tantalo et al. 1996, 1998a,b). A synthetic population has to be generated, containing sub-populations with different ages and metallicities. Then a synthetic spectrum must be generated for the mixed population and subsequently used as input for a synthetic, spectral fitting program to determine the underlying stellar populations. In this way one can establish the calibration of the spectrophotometric tool in a self-consistent way. Furthermore, an implicit verification can be made that the populations are consistent with those obtained from a CMD analysis with AMORE.
Acknowledgements
The research was partly supported by TMR grant ERBFMRX-CT96-0086 from the European Community (Network: Formation and Evolution of Galaxies), by the Italian Ministry of University, Scientific Research and Technology (MURST) and by the Italian Space Agency (ASI).
The data of the third test, in which one of the astrophysical parameters was fixed at its correct value as discussed in Sect. 4.3.3, are given in Table A.2. The data on the fourth test, in which one parameter was fixed one sigma from its correct value as discussed in Sect. 4.3.4, are given in Table A.3.
The tables in the Appendix are available in electronic form at http://www.edpsciences.org and at the CDS via anonymous ftp to cdsarc.u-strasbg.fr (130.79.128.5) or via http://cdsweb.u-strasbg.fr/cgi-bin/qcat?J/A+A/392/1129.
| model | pcross | rcross | rbrood | pcreep | pcorr | generation | |
| 1 | 0.85 | 1.00 | 1.00 | 0.0 | 0.0 | 0.17266 | 361 |
| 2 | 0.85 | 1.00 | 1.00 | 0.3 | 0.0 | 0.17266 | 361 |
| 3 | 0.85 | 1.00 | 1.00 | 0.7 | 0.0 | 0.17266 | 361 |
| 4 | 0.85 | 1.00 | 1.00 | 0.0 | 0.3 | 0.27551 | 221 |
| 5 | 0.85 | 1.00 | 1.00 | 0.0 | 0.7 | 0.26830 | 221 |
| 6 | 0.85 | 1.00 | 1.00 | 0.3 | 0.3 | 0.30488 | 381 |
| 7 | 0.85 | 1.00 | 1.00 | 0.7 | 0.3 | 0.32005 | 341 |
| 8 | 0.85 | 1.00 | 1.00 | 0.3 | 0.7 | 0.32167 | 201 |
| 9 | 0.85 | 1.00 | 1.00 | 0.7 | 0.7 | 0.38310 | 341 |
| 10 | 0.50 | 1.00 | 1.00 | 0.0 | 0.0 | 0.35662 | 381 |
| 11 | 0.50 | 1.00 | 1.00 | 0.3 | 0.0 | 0.35662 | 381 |
| 12 | 0.50 | 1.00 | 1.00 | 0.7 | 0.0 | 0.35662 | 381 |
| 13 | 0.50 | 1.00 | 1.00 | 0.0 | 0.3 | 0.34961 | 281 |
| 14 | 0.50 | 1.00 | 1.00 | 0.0 | 0.7 | 0.17008 | 261 |
| 15 | 0.50 | 1.00 | 1.00 | 0.3 | 0.3 | 0.36025 | 381 |
| 16 | 0.50 | 1.00 | 1.00 | 0.7 | 0.3 | 0.24048 | 261 |
| 17 | 0.50 | 1.00 | 1.00 | 0.3 | 0.7 | 0.23956 | 321 |
| 18 | 0.50 | 1.00 | 1.00 | 0.7 | 0.7 | 0.29463 | 321 |
| 19 | 0.85 | 2.00 | 1.00 | 0.0 | 0.0 | 0.31700 | 384 |
| 20 | 0.85 | 2.00 | 1.00 | 0.3 | 0.0 | 0.31700 | 384 |
| 21 | 0.85 | 2.00 | 1.00 | 0.7 | 0.0 | 0.31700 | 384 |
| 22 | 0.85 | 2.00 | 1.00 | 0.0 | 0.3 | 0.25740 | 341 |
| 23 | 0.85 | 2.00 | 1.00 | 0.0 | 0.7 | 0.35805 | 341 |
| 24 | 0.85 | 2.00 | 1.00 | 0.3 | 0.3 | 0.34120 | 181 |
| 25 | 0.85 | 2.00 | 1.00 | 0.7 | 0.3 | 0.31290 | 141 |
| 26 | 0.85 | 2.00 | 1.00 | 0.3 | 0.7 | 0.30913 | 341 |
| 27 | 0.85 | 2.00 | 1.00 | 0.7 | 0.7 | 0.32212 | 261 |
| 28 | 0.50 | 2.00 | 1.00 | 0.0 | 0.0 | 0.16122 | 261 |
| 29 | 0.50 | 2.00 | 1.00 | 0.3 | 0.0 | 0.16122 | 261 |
| 30 | 0.50 | 2.00 | 1.00 | 0.7 | 0.0 | 0.16122 | 261 |
| 31 | 0.50 | 2.00 | 1.00 | 0.0 | 0.3 | 0.36796 | 381 |
| 32 | 0.50 | 2.00 | 1.00 | 0.0 | 0.7 | 0.28494 | 381 |
| 33 | 0.50 | 2.00 | 1.00 | 0.3 | 0.3 | 0.27263 | 381 |
| 34 | 0.50 | 2.00 | 1.00 | 0.7 | 0.3 | 0.16352 | 301 |
| 35 | 0.50 | 2.00 | 1.00 | 0.3 | 0.7 | 0.35149 | 301 |
| 36 | 0.50 | 2.00 | 1.00 | 0.7 | 0.7 | 0.34031 | 381 |
| 37 | 0.85 | 3.00 | 1.00 | 0.0 | 0.0 | 0.38297 | 301 |
| 38 | 0.85 | 3.00 | 1.00 | 0.3 | 0.0 | 0.38297 | 301 |
| 39 | 0.85 | 3.00 | 1.00 | 0.7 | 0.0 | 0.38297 | 301 |
| 40 | 0.85 | 3.00 | 1.00 | 0.0 | 0.3 | 0.41019 | 341 |
| 41 | 0.85 | 3.00 | 1.00 | 0.0 | 0.7 | 0.32012 | 381 |
| 42 | 0.85 | 3.00 | 1.00 | 0.3 | 0.3 | 0.36840 | 341 |
| 43 | 0.85 | 3.00 | 1.00 | 0.7 | 0.3 | 0.31281 | 361 |
| 44 | 0.85 | 3.00 | 1.00 | 0.7 | 0.3 | 0.31281 | 361 |
| 45 | 0.85 | 3.00 | 1.00 | 0.3 | 0.7 | 0.30571 | 381 |
| 46 | 0.50 | 3.00 | 1.00 | 0.0 | 0.0 | 0.15614 | 301 |
| 47 | 0.50 | 3.00 | 1.00 | 0.3 | 0.0 | 0.15614 | 301 |
| 48 | 0.50 | 3.00 | 1.00 | 0.7 | 0.0 | 0.15614 | 301 |
| 49 | 0.50 | 3.00 | 1.00 | 0.0 | 0.3 | 0.23392 | 301 |
| 50 | 0.50 | 3.00 | 1.00 | 0.0 | 0.7 | 0.16274 | 221 |
| 51 | 0.50 | 3.00 | 1.00 | 0.3 | 0.3 | 0.25123 | 281 |
| 52 | 0.50 | 3.00 | 1.00 | 0.7 | 0.3 | 0.27151 | 261 |
| 53 | 0.50 | 3.00 | 1.00 | 0.3 | 0.7 | 0.35597 | 301 |
| model | pcross | rcross | rbrood | pcreep | pcorr | generation | |
| 54 | 0.50 | 3.00 | 1.00 | 0.7 | 0.7 | 0.27743 | 361 |
| 55 | 0.85 | 1.00 | 2.00 | 0.0 | 0.0 | 0.36221 | 161 |
| 56 | 0.85 | 1.00 | 2.00 | 0.3 | 0.0 | 0.36221 | 161 |
| 57 | 0.85 | 1.00 | 2.00 | 0.7 | 0.0 | 0.36221 | 161 |
| 58 | 0.85 | 1.00 | 2.00 | 0.0 | 0.3 | 0.16284 | 261 |
| 59 | 0.85 | 1.00 | 2.00 | 0.0 | 0.7 | 0.30251 | 381 |
| 60 | 0.85 | 1.00 | 2.00 | 0.3 | 0.3 | 0.32133 | 361 |
| 61 | 0.85 | 1.00 | 2.00 | 0.7 | 0.3 | 0.33565 | 361 |
| 62 | 0.85 | 1.00 | 2.00 | 0.3 | 0.7 | 0.27452 | 341 |
| 63 | 0.85 | 1.00 | 2.00 | 0.7 | 0.7 | 0.34660 | 301 |
| 64 | 0.50 | 1.00 | 2.00 | 0.0 | 0.0 | 0.27683 | 381 |
| 65 | 0.50 | 1.00 | 2.00 | 0.3 | 0.0 | 0.27683 | 381 |
| 66 | 0.50 | 1.00 | 2.00 | 0.7 | 0.0 | 0.27683 | 381 |
| 67 | 0.50 | 1.00 | 2.00 | 0.0 | 0.3 | 0.16833 | 201 |
| 68 | 0.50 | 1.00 | 2.00 | 0.0 | 0.7 | 0.38586 | 321 |
| 69 | 0.50 | 1.00 | 2.00 | 0.3 | 0.3 | 0.34390 | 361 |
| 70 | 0.50 | 1.00 | 2.00 | 0.7 | 0.3 | 0.33531 | 241 |
| 71 | 0.50 | 1.00 | 2.00 | 0.3 | 0.7 | 0.32646 | 221 |
| 72 | 0.50 | 1.00 | 2.00 | 0.7 | 0.7 | 0.34591 | 381 |
| 73 | 0.85 | 1.00 | 4.00 | 0.0 | 0.0 | 0.34607 | 221 |
| 74 | 0.85 | 1.00 | 4.00 | 0.3 | 0.0 | 0.34607 | 221 |
| 75 | 0.85 | 1.00 | 4.00 | 0.7 | 0.0 | 0.34607 | 221 |
| 76 | 0.85 | 1.00 | 4.00 | 0.0 | 0.3 | 0.32143 | 361 |
| 77 | 0.85 | 1.00 | 4.00 | 0.0 | 0.7 | 0.27159 | 381 |
| 78 | 0.85 | 1.00 | 4.00 | 0.3 | 0.3 | 0.26001 | 221 |
| 79 | 0.85 | 1.00 | 4.00 | 0.7 | 0.3 | 0.19802 | 241 |
| 80 | 0.85 | 1.00 | 4.00 | 0.3 | 0.7 | 0.33032 | 381 |
| 81 | 0.85 | 1.00 | 4.00 | 0.7 | 0.7 | 0.17799 | 341 |
| 82 | 0.50 | 1.00 | 4.00 | 0.0 | 0.0 | 0.27728 | 201 |
| 83 | 0.50 | 1.00 | 4.00 | 0.3 | 0.0 | 0.27728 | 201 |
| 84 | 0.50 | 1.00 | 4.00 | 0.7 | 0.0 | 0.27728 | 201 |
| 85 | 0.50 | 1.00 | 4.00 | 0.0 | 0.3 | 0.35783 | 381 |
| 86 | 0.50 | 1.00 | 4.00 | 0.0 | 0.7 | 0.28070 | 341 |
| 87 | 0.50 | 1.00 | 4.00 | 0.3 | 0.3 | 0.33717 | 381 |
| 88 | 0.50 | 1.00 | 4.00 | 0.7 | 0.3 | 0.18275 | 341 |
| 89 | 0.50 | 1.00 | 4.00 | 0.3 | 0.7 | 0.24795 | 361 |
| 90 | 0.50 | 1.00 | 4.00 | 0.7 | 0.7 | 0.28384 | 341 |
| 91 | 0.85 | 2.00 | 2.00 | 0.0 | 0.0 | 0.29274 | 261 |
| 92 | 0.85 | 2.00 | 2.00 | 0.3 | 0.0 | 0.29274 | 261 |
| 93 | 0.85 | 2.00 | 2.00 | 0.7 | 0.0 | 0.29274 | 261 |
| 94 | 0.85 | 2.00 | 2.00 | 0.0 | 0.3 | 0.24259 | 381 |
| 95 | 0.85 | 2.00 | 2.00 | 0.0 | 0.7 | 0.26513 | 161 |
| 96 | 0.85 | 2.00 | 2.00 | 0.3 | 0.3 | 0.33411 | 361 |
| 97 | 0.85 | 2.00 | 2.00 | 0.7 | 0.3 | 0.25808 | 121 |
| 98 | 0.85 | 2.00 | 2.00 | 0.3 | 0.7 | 0.35686 | 341 |
| 99 | 0.85 | 2.00 | 2.00 | 0.7 | 0.7 | 0.37254 | 399 |
| 100 | 0.50 | 2.00 | 2.00 | 0.0 | 0.0 | 0.28259 | 361 |
| 101 | 0.50 | 2.00 | 2.00 | 0.3 | 0.0 | 0.28259 | 361 |
| 102 | 0.50 | 2.00 | 2.00 | 0.7 | 0.0 | 0.28259 | 361 |
| 103 | 0.50 | 2.00 | 2.00 | 0.0 | 0.3 | 0.34559 | 341 |
| 104 | 0.50 | 2.00 | 2.00 | 0.0 | 0.7 | 0.39464 | 320 |
| 105 | 0.50 | 2.00 | 2.00 | 0.3 | 0.3 | 0.27112 | 181 |
| 106 | 0.50 | 2.00 | 2.00 | 0.7 | 0.3 | 0.29866 | 321 |
| 107 | 0.50 | 2.00 | 2.00 | 0.3 | 0.7 | 0.23660 | 181 |
| 108 | 0.50 | 2.00 | 2.00 | 0.7 | 0.7 | 0.35769 | 261 |
| 109 | 0.85 | 2.00 | 4.00 | 0.0 | 0.0 | 0.32628 | 181 |
| 110 | 0.85 | 2.00 | 4.00 | 0.3 | 0.0 | 0.32628 | 181 |
| 111 | 0.85 | 2.00 | 4.00 | 0.7 | 0.0 | 0.32628 | 181 |
| model | pcross | rcross | rbrood | pcreep | pcorr | generation | |
| 111 | 0.85 | 2.00 | 4.00 | 0.7 | 0.0 | 0.32628 | 181 |
| 112 | 0.85 | 2.00 | 4.00 | 0.0 | 0.3 | 0.26910 | 361 |
| 113 | 0.85 | 2.00 | 4.00 | 0.0 | 0.7 | 0.37278 | 389 |
| 114 | 0.85 | 2.00 | 4.00 | 0.3 | 0.3 | 0.31434 | 241 |
| 115 | 0.85 | 2.00 | 4.00 | 0.7 | 0.3 | 0.18222 | 321 |
| 116 | 0.85 | 2.00 | 4.00 | 0.3 | 0.7 | 0.26176 | 341 |
| 117 | 0.85 | 2.00 | 4.00 | 0.7 | 0.7 | 0.19495 | 241 |
| 118 | 0.50 | 2.00 | 4.00 | 0.0 | 0.0 | 0.17656 | 361 |
| 119 | 0.50 | 2.00 | 4.00 | 0.3 | 0.0 | 0.17656 | 361 |
| 120 | 0.50 | 2.00 | 4.00 | 0.7 | 0.0 | 0.17656 | 361 |
| 121 | 0.50 | 2.00 | 4.00 | 0.0 | 0.3 | 0.17248 | 181 |
| 122 | 0.50 | 2.00 | 4.00 | 0.0 | 0.7 | 0.27790 | 381 |
| 123 | 0.50 | 2.00 | 4.00 | 0.3 | 0.3 | 0.17117 | 221 |
| 124 | 0.50 | 2.00 | 4.00 | 0.7 | 0.3 | 0.36171 | 381 |
| 125 | 0.50 | 2.00 | 4.00 | 0.3 | 0.7 | 0.38431 | 392 |
| 126 | 0.50 | 2.00 | 4.00 | 0.7 | 0.7 | 0.31711 | 400 |
| 127 | 0.85 | 3.00 | 2.00 | 0.0 | 0.0 | 0.16288 | 161 |
| 128 | 0.85 | 3.00 | 2.00 | 0.3 | 0.0 | 0.16288 | 161 |
| 129 | 0.85 | 3.00 | 2.00 | 0.7 | 0.0 | 0.16288 | 161 |
| 130 | 0.85 | 3.00 | 2.00 | 0.0 | 0.3 | 0.34149 | 341 |
| 131 | 0.85 | 3.00 | 2.00 | 0.0 | 0.7 | 0.33630 | 281 |
| 132 | 0.85 | 3.00 | 2.00 | 0.3 | 0.3 | 0.24355 | 301 |
| 133 | 0.85 | 3.00 | 2.00 | 0.7 | 0.3 | 0.33731 | 221 |
| 134 | 0.85 | 3.00 | 2.00 | 0.3 | 0.7 | 0.26283 | 201 |
| 135 | 0.85 | 3.00 | 2.00 | 0.7 | 0.7 | 0.25975 | 121 |
| 136 | 0.50 | 3.00 | 2.00 | 0.0 | 0.0 | 0.26047 | 221 |
| 137 | 0.50 | 3.00 | 2.00 | 0.3 | 0.0 | 0.26047 | 221 |
| 138 | 0.50 | 3.00 | 2.00 | 0.7 | 0.0 | 0.26047 | 221 |
| 139 | 0.50 | 3.00 | 2.00 | 0.0 | 0.3 | 0.26449 | 321 |
| 140 | 0.50 | 3.00 | 2.00 | 0.0 | 0.7 | 0.27116 | 281 |
| 141 | 0.50 | 3.00 | 2.00 | 0.3 | 0.3 | 0.31298 | 101 |
| 142 | 0.50 | 3.00 | 2.00 | 0.7 | 0.3 | 0.32952 | 201 |
| 143 | 0.50 | 3.00 | 2.00 | 0.3 | 0.7 | 0.25496 | 281 |
| 144 | 0.50 | 3.00 | 2.00 | 0.7 | 0.7 | 0.24888 | 221 |
| 145 | 0.85 | 3.00 | 4.00 | 0.0 | 0.0 | 0.34932 | 321 |
| 146 | 0.85 | 3.00 | 4.00 | 0.3 | 0.0 | 0.34932 | 321 |
| 147 | 0.85 | 3.00 | 4.00 | 0.7 | 0.0 | 0.34932 | 321 |
| 148 | 0.85 | 3.00 | 4.00 | 0.0 | 0.3 | 0.34687 | 381 |
| 149 | 0.85 | 3.00 | 4.00 | 0.0 | 0.7 | 0.32433 | 321 |
| 150 | 0.85 | 3.00 | 4.00 | 0.3 | 0.3 | 0.17152 | 141 |
| 151 | 0.85 | 3.00 | 4.00 | 0.7 | 0.3 | 0.30958 | 395 |
| 152 | 0.85 | 3.00 | 4.00 | 0.3 | 0.7 | 0.31341 | 398 |
| 153 | 0.85 | 3.00 | 4.00 | 0.7 | 0.7 | 0.27072 | 261 |
| 154 | 0.50 | 3.00 | 4.00 | 0.0 | 0.0 | 0.34272 | 101 |
| 155 | 0.50 | 3.00 | 4.00 | 0.3 | 0.0 | 0.34272 | 101 |
| 156 | 0.50 | 3.00 | 4.00 | 0.7 | 0.0 | 0.34272 | 101 |
| 157 | 0.50 | 3.00 | 4.00 | 0.0 | 0.3 | 0.30844 | 321 |
| 158 | 0.50 | 3.00 | 4.00 | 0.0 | 0.7 | 0.27009 | 361 |
| 159 | 0.50 | 3.00 | 4.00 | 0.3 | 0.3 | 0.17333 | 381 |
| 160 | 0.50 | 3.00 | 4.00 | 0.7 | 0.3 | 0.25621 | 361 |
| 161 | 0.50 | 3.00 | 4.00 | 0.3 | 0.7 | 0.39429 | 341 |
| 162 | 0.50 | 3.00 | 4.00 | 0.7 | 0.7 | 0.26150 | 361 |
| parameter | model | log d (pc) |
|
|
|
|
||||
| log d | 9 | 0.34897 | 3.90633 | 0.002 | 9.89178 | 9.95342 | - 0.60089 | 0.19300 | 2.341 | 0.985 |
| 14 | 0.35919 | 3.90633 | - 0.001 | 9.88937 | 9.95245 | - 0.60115 | 0.20684 | 2.341 | 1.111 | |
| 22 | 0.26617 | 3.90633 | 0.007 | 9.86685 | 9.95757 | - 0.53389 | 0.30054 | 2.351 | 2.980 | |
| 34 | 0.31373 | 3.90633 | 0.001 | 9.87873 | 9.95335 | - 0.57392 | 0.25198 | 2.341 | 1.613 | |
| 40 | 0.15102 | 3.90633 | 0.016 | 9.78469 | 10.12246 | - 0.69199 | 0.07931 | 2.319 | - 1.281 | |
| 52 | 0.35475 | 3.90633 | 0.012 | 9.89392 | 9.95002 | - 0.59523 | 0.17305 | 2.338 | 0.936 | |
| 9 | 0.25192 | 3.89083 | 0.000 | 9.87519 | 9.98656 | - 0.50206 | 0.34423 | 2.391 | 2.613 | |
| 14 | 0.23362 | 3.88471 | 0.000 | 9.88175 | 9.99802 | - 0.50635 | 0.40461 | 2.424 | 3.682 | |
| 22 | 0.30930 | 3.89399 | - 0.001 | 9.91339 | 9.98285 | - 0.55292 | 0.20319 | 2.383 | 0.889 | |
| 34 | 0.17545 | 3.89688 | 0.001 | 9.80693 | 10.1395 | - 0.66073 | 0.10056 | 2.339 | - 1.338 | |
| 40 | 0.26131 | 3.89634 | 0.001 | 9.82335 | 9.96459 | - 0.51903 | 0.44994 | 2.355 | 3.493 | |
| 52 | 0.41406 | 3.90592 | 0.000 | 9.90287 | 9.95544 | - 0.59621 | 0.17710 | 2.350 | 1.003 | |
| log
|
9 | 0.34505 | 3.89687 | 0.030 | 9.90310 | 9.96292 | - 0.59785 | 0.19586 | 2.358 | 1.028 |
| 14 | 0.29609 | 3.90078 | 0.064 | 9.90308 | 9.93943 | - 0.67152 | 0.10196 | 2.333 | 0.087 | |
| 22 | 0.34452 | 3.89289 | 0.026 | 9.90308 | 9.96482 | - 0.57260 | 0.23664 | 2.362 | 1.435 | |
| 34 | 0.32855 | 3.89861 | 0.067 | 9.90309 | 9.94303 | - 0.62670 | 0.14375 | 2.343 | 0.845 | |
| 40 | 0.35429 | 3.89657 | 0.014 | 9.90309 | 9.96685 | - 0.56333 | 0.21007 | 2.358 | 1.130 | |
| 52 | 0.32296 | 3.89388 | 0.036 | 9.90309 | 9.96827 | - 0.58340 | 0.21130 | 2.365 | 1.197 | |
| log
|
9 | 0.32414 | 3.89839 | 0.033 | 9.88329 | 9.95424 | - 0.55256 | 0.26193 | 2.341 | 1.674 |
| 14 | 0.25502 | 3.89455 | 0.036 | 9.85431 | 9.95425 | - 0.52027 | 0.39343 | 2.351 | 3.455 | |
| 22 | 0.26055 | 3.90038 | 0.022 | 9.76480 | 9.95423 | - 0.52358 | 0.52141 | 2.333 | 4.267 | |
| 34 | 0.36575 | 3.89772 | 0.046 | 9.90052 | 9.95424 | - 0.60081 | 0.20274 | 2.352 | 1.251 | |
| 40 | 0.25141 | 3.89839 | 0.042 | 9.75973 | 9.95424 | - 0.56527 | 0.52340 | 2.335 | 4.878 | |
| 52 | 0.37074 | 3.89458 | 0.066 | 9.91101 | 9.95424 | - 0.62634 | 0.16462 | 2.361 | 1.046 | |
| log
|
9 | 0.22556 | 3.89237 | 0.059 | 9.78605 | 9.96444 | - 0.60205 | 0.48867 | 2.363 | 4.119 |
| 14 | 0.32714 | 3.89352 | 0.031 | 9.89643 | 9.96409 | - 0.60207 | 0.22149 | 2.354 | 0.907 | |
| 22 | 0.23902 | 3.89510 | 0.047 | 9.82960 | 9.95708 | - 0.60205 | 0.45203 | 2.363 | 3.709 | |
| 34 | 0.27029 | 3.89729 | 0.040 | 9.84880 | 9.95803 | - 0.60207 | 0.31992 | 2.327 | 1.756 | |
| 40 | 0.28851 | 3.89175 | 0.028 | 9.94114 | 9.97064 | - 0.60206 | 0.10911 | 2.399 | 0.305 | |
| 52 | 0.26238 | 3.89419 | 0.041 | 9.87970 | 9.96558 | - 0.60207 | 0.29439 | 2.375 | 2.197 | |
| log
|
9 | 0.32971 | 3.89633 | 0.044 | 9.90542 | 9.96035 | - 0.58239 | 0.17609 | 2.363 | 1.182 |
| 14 | 0.39924 | 3.89593 | 0.038 | 9.90880 | 9.96147 | - 0.58144 | 0.17608 | 2.360 | 1.009 | |
| 22 | 0.40156 | 3.89579 | 0.053 | 9.90555 | 9.95702 | - 0.60422 | 0.17608 | 2.357 | 1.057 | |
| 34 | 0.31199 | 3.89306 | 0.017 | 9.92288 | 9.97460 | - 0.57681 | 0.17610 | 2.384 | 0.872 | |
| 40 | 0.35466 | 3.89952 | 0.044 | 9.90467 | 9.95065 | - 0.61730 | 0.17609 | 2.349 | 1.010 | |
| 52 | 0.36582 | 3.89853 | 0.042 | 9.90077 | 9.95166 | - 0.60430 | 0.17608 | 2.342 | 0.878 | |
| 9 | 0.34047 | 3.89758 | 0.023 | 9.89805 | 9.96360 | - 0.57367 | 0.19013 | 2.350 | 0.831 | |
| 14 | 0.17505 | 3.89826 | 0.014 | 9.82400 | 10.14663 | - 0.62474 | 0.04615 | 2.349 | - 1.950 | |
| 22 | 0.30768 | 3.89446 | 0.063 | 9.90882 | 9.95914 | - 0.73991 | 0.12715 | 2.349 | - 0.035 | |
| 34 | 0.31327 | 3.89614 | 0.019 | 9.87877 | 9.96638 | - 0.52373 | 0.26868 | 2.350 | 1.784 | |
| 40 | 0.35728 | 3.89731 | 0.021 | 9.89219 | 9.96144 | - 0.55927 | 0.23623 | 2.349 | 1.347 | |
| 52 | 0.33530 | 3.89702 | 0.027 | 9.89860 | 9.96481 | - 0.54426 | 0.19782 | 2.350 | 1.039 | |
| 9 | 0.31038 | 3.89352 | 0.082 | 9.91755 | 9.94928 | - 0.62624 | 0.13301 | 2.360 | 1.000 | |
| 14 | 0.38179 | 3.89881 | 0.033 | 9.90224 | 9.95851 | - 0.57609 | 0.17513 | 2.352 | 1.000 | |
| 22 | 0.34095 | 3.89848 | 0.052 | 9.89348 | 9.95316 | - 0.57848 | 0.18922 | 2.338 | 0.999 | |
| 34 | 0.16612 | 3.89537 | 0.028 | 9.80857 | 10.0490 | - 0.44888 | 0.12520 | 2.382 | 1.001 | |
| 40 | 0.35606 | 3.90903 | 0.019 | 9.88940 | 9.94183 | - 0.61022 | 0.17903 | 2.329 | 1.001 | |
| 52 | 0.31834 | 3.89857 | 0.073 | 9.91742 | 9.93856 | - 0.61263 | 0.12154 | 2.350 | 1.000 |
| parameter | offset | model | log d |
|
|
|
|
||||
| log d | 9 | 0.21045 | 3.90307 | 0.008 | 9.70661 | 9.95123 | - 0.54348 | 0.59806 | 2.318 | 4.573 | |
| 14 | 0.29182 | 3.90307 | 0.005 | 9.92758 | 9.95781 | - 0.64641 | 0.10745 | 2.368 | 0.062 | ||
| 22 | 0.30663 | 3.90307 | 0.055 | 9.90867 | 9.93700 | - 0.62003 | 0.13004 | 2.332 | 0.373 | ||
| 34 | 0.27406 | 3.90307 | 0.009 | 9.93190 | 9.95224 | - 0.64172 | 0.09083 | 2.376 | 0.329 | ||
| 40 | 0.30508 | 3.90307 | 0.031 | 9.88057 | 9.94765 | - 0.56412 | 0.23885 | 2.330 | 1.482 | ||
| 52 | 0.35736 | 3.90307 | 0.051 | 9.89002 | 9.94201 | - 0.61187 | 0.18455 | 2.331 | 1.072 | ||
| 9 | 0.29273 | 3.90960 | 0.028 | 9.90776 | 9.93160 | - 0.64116 | 0.10185 | 2.333 | 0.378 | ||
| 14 | 0.33350 | 3.90960 | 0.007 | 9.89160 | 9.94261 | - 0.62176 | 0.20013 | 2.336 | 1.201 | ||
| 22 | 0.29174 | 3.90960 | 0.017 | 9.90814 | 9.94062 | - 0.63653 | 0.11803 | 2.337 | 0.279 | ||
| 34 | 0.15593 | 3.90960 | 0.000 | 9.82433 | 10.13280 | - 0.74638 | 0.00763 | 2.346 | - 1.894 | ||
| 40 | 0.33700 | 3.90960 | 0.008 | 9.89589 | 9.94669 | - 0.60483 | 0.15343 | 2.335 | 0.771 | ||
| 52 | 0.26929 | 3.90960 | 0.032 | 9.91164 | 9.93459 | - 0.64348 | 0.07215 | 2.340 | 0.145 | ||
| 9 | - | - | - | - | - | - | - | - | - | ||
| 14 | 0.27049 | 3.89442 | 0.014 | 9.79866 | 9.96608 | - 0.52682 | 0.49361 | 2.358 | 4.064 | ||
| 22 | 0.28352 | 3.89378 | 0.014 | 9.87464 | 9.97332 | - 0.55944 | 0.30339 | 2.359 | 1.855 | ||
| 34 | 0.28608 | 3.89345 | 0.014 | 9.88671 | 9.97642 | - 0.54707 | 0.30071 | 2.376 | 1.973 | ||
| 40 | 0.26382 | 3.89346 | 0.014 | 9.81948 | 9.96660 | - 0.52576 | 0.46184 | 2.358 | 3.610 | ||
| 52 | 0.31545 | 3.89363 | 0.014 | 9.90003 | 9.97313 | - 0.56649 | 0.23400 | 2.372 | 1.349 | ||
| log
|
9 | 0.27236 | 3.90040 | 0.040 | 9.85386 | 9.94757 | - 0.55836 | 0.30935 | 2.323 | 1.896 | |
| 14 | 0.27545 | 3.89466 | 0.019 | 9.85386 | 9.97024 | - 0.52819 | 0.37310 | 2.368 | 2.888 | ||
| 22 | 0.27529 | 3.89648 | 0.018 | 9.85386 | 9.96558 | - 0.54638 | 0.35495 | 2.350 | 2.384 | ||
| 34 | 0.16883 | 3.89375 | 0.029 | 9.85386 | 10.08302 | - 0.47457 | 0.01306 | 2.400 | - 0.280 | ||
| 40 | 0.28900 | 3.89953 | 0.033 | 9.85386 | 9.95405 | - 0.56448 | 0.30320 | 2.324 | 1.779 | ||
| 52 | 0.26942 | 3.89372 | 0.047 | 9.85386 | 9.96388 | - 0.55833 | 0.34280 | 2.356 | 2.939 | ||
| 9 | 0.26839 | 3.89058 | 0.041 | 9.95232 | 9.95626 | - 0.54291 | 0.11856 | 2.399 | 0.517 | ||
| 14 | 0.27352 | 3.88923 | 0.037 | 9.95232 | 9.96263 | - 0.68969 | 0.08041 | 2.401 | - 0.347 | ||
| 22 | 0.27555 | 3.88867 | 0.037 | 9.95232 | 9.95915 | - 0.59030 | 0.06926 | 2.399 | - 0.175 | ||
| 34 | 0.29222 | 3.88687 | 0.054 | 9.95232 | 9.95543 | - 0.60299 | 0.07026 | 2.398 | - 0.115 | ||
| 40 | 0.26972 | 3.88983 | 0.045 | 9.95232 | 9.95314 | - 0.61176 | 0.08503 | 2.400 | 0.007 | ||
| 52 | 0.29660 | 3.89064 | 0.046 | 9.95232 | 9.95592 | - 0.65754 | 0.08096 | 2.401 | 0.003 | ||
| log
|
9 | 0.21567 | 3.90212 | 0.093 | 9.90420 | 9.90749 | - 0.59379 | 0.10497 | 2.306 | 0.206 | |
| 14 | 0.25127 | 3.90319 | 0.111 | 9.90723 | 9.90749 | - 0.68463 | 0.09099 | 2.318 | 0.272 | ||
| 22 | 0.23271 | 3.90019 | 0.114 | 9.90634 | 9.90749 | - 0.69157 | 0.08733 | 2.316 | 0.167 | ||
| 34 | 0.24395 | 3.90526 | 0.113 | 9.89882 | 9.90749 | - 0.64179 | 0.09370 | 2.307 | 0.470 | ||
| 40 | 0.23796 | 3.90273 | 0.106 | 9.90270 | 9.90750 | - 0.63701 | 0.10850 | 2.306 | 0.378 | ||
| 52 | 0.19091 | 3.90474 | 0.105 | 9.85263 | 9.90750 | - 0.54484 | 0.26544 | 2.274 | 2.304 | ||
| 9 | 0.18966 | 3.88133 | - 0.001 | 9.84694 | 10.00986 | - 0.51375 | 0.40765 | 2.432 | 3.306 | ||
| 14 | 0.22621 | 3.88298 | - 0.001 | 9.91537 | 10.00986 | - 0.50616 | 0.26567 | 2.437 | 2.059 | ||
| 22 | 0.21599 | 3.88275 | 0.000 | 9.73977 | 10.00986 | - 0.51424 | 0.51570 | 2.413 | 4.449 | ||
| 34 | 0.20975 | 3.88419 | - 0.001 | 9.91194 | 10.00985 | - 0.51870 | 0.26155 | 2.427 | 2.046 | ||
| 40 | 0.19191 | 3.88460 | 0.013 | 9.79535 | 10.00985 | - 0.52199 | 0.43616 | 2.405 | 3.653 | ||
| 52 | 0.21132 | 3.88557 | 0.002 | 9.90671 | 10.00987 | - 0.51864 | 0.27274 | 2.422 | 2.044 | ||
| log
|
9 | 0.32236 | 3.89424 | 0.060 | 9.92490 | 9.94949 | - 0.65235 | 0.09935 | 2.364 | 0.023 | |
| 14 | 0.19541 | 3.89205 | 0.085 | 9.80620 | 9.95304 | - 0.65235 | 0.44787 | 2.367 | 4.219 | ||
| 22 | 0.19646 | 3.89803 | 0.068 | 9.80556 | 9.95382 | - 0.65235 | 0.48659 | 2.368 | 4.525 | ||
| 34 | 0.19203 | 3.89730 | 0.066 | 9.76582 | 9.94810 | - 0.65235 | 0.48742 | 2.316 | 4.023 | ||
| 40 | 0.32867 | 3.89484 | 0.038 | 9.90919 | 9.95602 | - 0.65235 | 0.18180 | 2.356 | 0.601 | ||
| 52 | 0.28939 | 3.89843 | 0.035 | 9.88752 | 9.95511 | - 0.65235 | 0.22274 | 2.337 | 0.836 | ||
| 9 | 0.30963 | 3.89302 | 0.019 | 9.89547 | 9.97009 | - 0.55177 | 0.26207 | 2.372 | 1.833 | ||
| 14 | 0.32883 | 3.89295 | 0.022 | 9.91000 | 9.97315 | - 0.55177 | 0.19130 | 2.365 | 0.778 | ||
| 22 | 0.32293 | 3.89110 | 0.024 | 9.91831 | 9.96435 | - 0.55177 | 0.18106 | 2.369 | 0.671 | ||
| 34 | 0.26376 | 3.89625 | 0.024 | 9.86434 | 9.96369 | - 0.55177 | 0.37255 | 2.361 | 2.928 | ||
| 40 | 0.33593 | 3.89840 | 0.032 | 9.90057 | 9.96348 | - 0.55177 | 0.17853 | 2.349 | 0.885 | ||
| 52 | 0.32988 | 3.89341 | 0.033 | 9.90827 | 9.97162 | - 0.55177 | 0.20534 | 2.376 | 1.416 | ||
| log
|
9 | 0.29612 | 3.89604 | 0.077 | 9.92807 | 9.93582 | - 0.66338 | 0.04239 | 2.356 | - 0.232 | |
| 14 | 0.29094 | 3.89708 | 0.082 | 9.92897 | 9.93400 | - 0.67421 | 0.04239 | 2.359 | - 0.139 | ||
| 22 | 0.27692 | 3.89364 | 0.066 | 9.94038 | 9.95069 | - 0.66032 | 0.04239 | 2.387 | - 0.337 | ||
| 34 | 0.28980 | 3.89736 | 0.083 | 9.92808 | 9.93087 | - 0.63329 | 0.04239 | 2.357 | - 0.095 | ||
| 40 | 0.16160 | 3.90196 | 0.043 | 9.82928 | 10.10576 | - 0.79270 | 0.04238 | 2.333 | - 1.676 | ||
| 52 | 0.30348 | 3.89643 | 0.078 | 9.92740 | 9.93508 | - 0.66677 | 0.04239 | 2.355 | - 0.238 | ||
| 9 | 0.26702 | 3.88982 | 0.031 | 9.88500 | 9.97615 | - 0.56217 | 0.30979 | 2.384 | 2.280 | ||
| 14 | 0.29357 | 3.89392 | 0.012 | 9.88192 | 9.97494 | - 0.53776 | 0.30979 | 2.371 | 2.129 | ||
| 22 | 0.30635 | 3.89366 | 0.032 | 9.87176 | 9.96383 | - 0.53473 | 0.30979 | 2.351 | 2.194 | ||
| 34 | 0.25823 | 3.89389 | 0.046 | 9.87585 | 9.96245 | - 0.53400 | 0.30979 | 2.357 | 2.578 | ||
| 40 | 0.28417 | 3.89886 | 0.028 | 9.85863 | 9.95559 | - 0.57966 | 0.30979 | 2.333 | 1.926 | ||
| 52 | 0.28883 | 3.89501 | 0.020 | 9.88098 | 9.96677 | - 0.55136 | 0.30979 | 2.367 | 2.284 | ||
| 9 | 0.24524 | 3.90004 | 0.053 | 9.85726 | 9.94126 | - 0.56638 | 0.28025 | 2.316 | 1.932 | ||
| 14 | 0.28147 | 3.90272 | 0.020 | 9.87188 | 9.94707 | - 0.54698 | 0.24944 | 2.316 | 1.230 | ||
| 22 | 0.27797 | 3.89896 | 0.042 | 9.86368 | 9.94580 | - 0.54982 | 0.26102 | 2.316 | 1.511 | ||
| 34 | 0.22427 | 3.89644 | 0.032 | 9.82350 | 9.94534 | - 0.50532 | 0.45532 | 2.316 | 3.679 | ||
| 40 | 0.27857 | 3.90056 | 0.042 | 9.86375 | 9.94608 | - 0.59271 | 0.26364 | 2.316 | 1.294 | ||
| 52 | 0.28640 | 3.90139 | 0.039 | 9.85871 | 9.94768 | - 0.58962 | 0.27721 | 2.316 | 1.438 | ||
| 9 | 0.23946 | 3.89295 | 0.062 | 9.87606 | 9.96018 | - 0.57093 | 0.35127 | 2.384 | 3.499 | ||
| 14 | 0.25586 | 3.89172 | 0.014 | 9.86873 | 9.97973 | - 0.53518 | 0.35320 | 2.384 | 2.919 | ||
| 22 | 0.28357 | 3.89109 | 0.021 | 9.91357 | 9.97110 | - 0.56888 | 0.24524 | 2.384 | 1.488 | ||
| 34 | 0.29934 | 3.89146 | 0.023 | 9.91497 | 9.97387 | - 0.58765 | 0.23086 | 2.384 | 1.461 | ||
| 40 | 0.25505 | 3.89153 | 0.009 | 9.80529 | 9.97968 | - 0.52454 | 0.51555 | 2.384 | 4.820 | ||
| 52 | 0.30437 | 3.89128 | 0.026 | 9.90426 | 9.97836 | - 0.55141 | 0.23875 | 2.384 | 1.620 |
| parameter | offset | model | log d |
|
|
|
|
||||
|
|
9 | 0.29110 | 3.89717 | 0.083 | 9.93098 | 9.93961 | - 0.68243 | 0.02188 | 2.365 | - 0.397 | |
| 14 | 0.17266 | 3.89314 | 0.001 | 9.83767 | 10.08181 | - 0.41912 | 0.06629 | 2.372 | - 0.397 | ||
| 22 | 0.30211 | 3.89619 | 0.084 | 9.91729 | 9.94002 | - 0.68522 | 0.06730 | 2.348 | - 0.397 | ||
| 34 | 0.16366 | 3.89491 | 0.015 | 9.81889 | 10.08661 | - 0.50001 | 0.09684 | 2.363 | - 0.397 | ||
| 40 | 0.16472 | 3.89974 | 0.018 | 9.80958 | 10.07264 | - 0.54158 | 0.11992 | 2.357 | - 0.397 | ||
| 52 | 0.29684 | 3.89592 | 0.074 | 9.91808 | 9.93138 | - 0.66681 | 0.08493 | 2.339 | - 0.397 | ||
| 9 | 0.26953 | 3.89638 | 0.045 | 9.86719 | 9.95699 | - 0.57054 | 0.30738 | 2.346 | 2.397 | ||
| 14 | 0.29111 | 3.89398 | 0.023 | 9.87948 | 9.97371 | - 0.54141 | 0.30286 | 2.371 | 2.397 | ||
| 22 | 0.28842 | 3.89548 | 0.029 | 9.86193 | 9.95692 | - 0.53941 | 0.33240 | 2.341 | 2.397 | ||
| 34 | 0.27336 | 3.89607 | 0.048 | 9.88719 | 9.95851 | - 0.56455 | 0.27131 | 2.355 | 2.397 | ||
| 40 | 0.26195 | 3.89766 | 0.038 | 9.87053 | 9.94793 | - 0.58042 | 0.30908 | 2.345 | 2.397 | ||
| 52 | 0.27328 | 3.89034 | 0.022 | 9.87427 | 9.97430 | - 0.54963 | 0.33427 | 2.375 | 2.397 |